Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
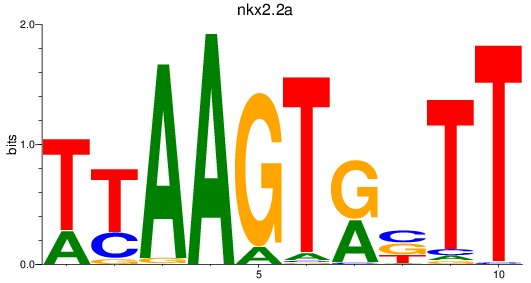
Results for nkx2.2a
Z-value: 1.11
Transcription factors associated with nkx2.2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nkx2.2a
|
ENSDARG00000053298 | NK2 homeobox 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nkx2.2a | dr11_v1_chr17_-_42218652_42218652 | -0.89 | 1.4e-03 | Click! |
Activity profile of nkx2.2a motif
Sorted Z-values of nkx2.2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_-_13690712 | 2.50 |
ENSDART00000065817
|
pou5f3
|
POU domain, class 5, transcription factor 3 |
| chr24_-_33308045 | 2.30 |
ENSDART00000149711
|
slc4a2b
|
solute carrier family 4 (anion exchanger), member 2b |
| chr2_-_38992304 | 2.02 |
ENSDART00000114085
ENSDART00000146812 |
si:ch211-119o8.6
|
si:ch211-119o8.6 |
| chr11_+_42422371 | 1.85 |
ENSDART00000163780
|
dennd6aa
|
DENN/MADD domain containing 6Aa |
| chr11_+_42422638 | 1.75 |
ENSDART00000042599
ENSDART00000181175 |
dennd6aa
|
DENN/MADD domain containing 6Aa |
| chr23_-_27045231 | 1.75 |
ENSDART00000187979
|
zgc:66440
|
zgc:66440 |
| chr17_+_6538733 | 1.65 |
ENSDART00000193712
|
slc5a6b
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chr13_+_47710434 | 1.63 |
ENSDART00000188724
|
TMEM87B
|
transmembrane protein 87B |
| chr8_+_52442622 | 1.60 |
ENSDART00000012758
|
zgc:77112
|
zgc:77112 |
| chr16_-_54919260 | 1.51 |
ENSDART00000156533
|
kdf1a
|
keratinocyte differentiation factor 1a |
| chr2_-_15318786 | 1.47 |
ENSDART00000135851
|
tecrl2b
|
trans-2,3-enoyl-CoA reductase-like 2b |
| chr2_-_15324837 | 1.44 |
ENSDART00000015655
|
tecrl2b
|
trans-2,3-enoyl-CoA reductase-like 2b |
| chr15_+_29025090 | 1.40 |
ENSDART00000131755
|
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr2_+_7557912 | 1.39 |
ENSDART00000160053
|
ripk2
|
receptor-interacting serine-threonine kinase 2 |
| chr18_-_43866001 | 1.38 |
ENSDART00000150218
|
treh
|
trehalase (brush-border membrane glycoprotein) |
| chr8_-_1698155 | 1.32 |
ENSDART00000186159
|
CABZ01065417.1
|
|
| chr22_+_25249193 | 1.31 |
ENSDART00000171851
|
si:ch211-226h8.11
|
si:ch211-226h8.11 |
| chr1_-_55248496 | 1.24 |
ENSDART00000098615
|
nanos3
|
nanos homolog 3 |
| chr20_-_34750045 | 1.23 |
ENSDART00000186130
|
znf395b
|
zinc finger protein 395b |
| chr12_-_6880694 | 1.21 |
ENSDART00000171846
|
pcdh15b
|
protocadherin-related 15b |
| chr14_-_24110707 | 1.21 |
ENSDART00000133522
ENSDART00000123152 |
cpeb4a
|
cytoplasmic polyadenylation element binding protein 4a |
| chr22_+_25236657 | 1.21 |
ENSDART00000138012
|
zgc:172218
|
zgc:172218 |
| chr11_+_18157260 | 1.12 |
ENSDART00000144659
|
zgc:173545
|
zgc:173545 |
| chr1_+_30723380 | 1.12 |
ENSDART00000127943
ENSDART00000062628 ENSDART00000127670 |
bora
|
bora, aurora kinase A activator |
| chr23_-_28120058 | 1.11 |
ENSDART00000087815
|
b4galnt1a
|
beta-1,4-N-acetyl-galactosaminyl transferase 1a |
| chr25_+_35375848 | 1.08 |
ENSDART00000155721
|
ano3
|
anoctamin 3 |
| chr19_-_27827744 | 1.08 |
ENSDART00000181620
|
papd7
|
PAP associated domain containing 7 |
| chr18_+_26428829 | 1.06 |
ENSDART00000190779
ENSDART00000193226 ENSDART00000110746 |
blm
|
Bloom syndrome, RecQ helicase-like |
| chr11_+_37638873 | 1.06 |
ENSDART00000186384
ENSDART00000184291 ENSDART00000131782 ENSDART00000140502 |
sh2d5
|
SH2 domain containing 5 |
| chr24_+_15655069 | 1.06 |
ENSDART00000042943
ENSDART00000181296 |
fbxo15
|
F-box protein 15 |
| chr24_+_41989108 | 1.06 |
ENSDART00000169725
|
zbtb14
|
zinc finger and BTB domain containing 14 |
| chr19_+_34230108 | 1.04 |
ENSDART00000141950
|
galnt12
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 12 |
| chr16_-_28593951 | 1.03 |
ENSDART00000183322
|
rpp38
|
ribonuclease P/MRP 38 subunit |
| chr9_+_2762270 | 1.01 |
ENSDART00000123342
ENSDART00000001795 ENSDART00000177563 |
sp3a
|
sp3a transcription factor |
| chr12_-_33789006 | 1.00 |
ENSDART00000034550
|
llgl2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr12_-_33789218 | 0.99 |
ENSDART00000193258
|
llgl2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr21_+_19547806 | 0.99 |
ENSDART00000159707
ENSDART00000184869 ENSDART00000181321 ENSDART00000058487 ENSDART00000058485 |
rai14
|
retinoic acid induced 14 |
| chr10_+_585719 | 0.99 |
ENSDART00000180167
|
smad4a
|
SMAD family member 4a |
| chr13_-_30996072 | 0.98 |
ENSDART00000181661
|
wdfy4
|
WDFY family member 4 |
| chr1_-_40341306 | 0.96 |
ENSDART00000190649
|
maml3
|
mastermind-like transcriptional coactivator 3 |
| chr14_+_29200772 | 0.95 |
ENSDART00000166608
|
TENM2
|
si:dkey-34l15.2 |
| chr2_+_50477779 | 0.94 |
ENSDART00000122716
|
CABZ01067973.1
|
|
| chr17_+_24809221 | 0.94 |
ENSDART00000082251
ENSDART00000147871 ENSDART00000130871 |
spdya
|
speedy/RINGO cell cycle regulator family member A |
| chr23_+_38245610 | 0.93 |
ENSDART00000191386
|
znf217
|
zinc finger protein 217 |
| chr17_+_24809743 | 0.93 |
ENSDART00000135986
|
spdya
|
speedy/RINGO cell cycle regulator family member A |
| chr5_-_16472719 | 0.92 |
ENSDART00000162071
|
piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr5_-_37117778 | 0.92 |
ENSDART00000149138
|
il13ra2
|
interleukin 13 receptor, alpha 2 |
| chr10_-_32851847 | 0.90 |
ENSDART00000134255
|
trim37
|
tripartite motif containing 37 |
| chr1_+_8534698 | 0.90 |
ENSDART00000021504
|
smcr8b
|
Smith-Magenis syndrome chromosome region, candidate 8b |
| chr5_+_16117871 | 0.89 |
ENSDART00000090657
|
znrf3
|
zinc and ring finger 3 |
| chr15_-_44601331 | 0.89 |
ENSDART00000161514
|
zgc:165508
|
zgc:165508 |
| chr3_-_48603471 | 0.89 |
ENSDART00000189027
|
ndel1b
|
nudE neurodevelopment protein 1-like 1b |
| chr9_+_28140089 | 0.89 |
ENSDART00000046880
|
plekhm3
|
pleckstrin homology domain containing, family M, member 3 |
| chr20_-_35512932 | 0.88 |
ENSDART00000137690
|
adgrf3b
|
adhesion G protein-coupled receptor F3b |
| chr10_+_36441124 | 0.88 |
ENSDART00000185626
|
uspl1
|
ubiquitin specific peptidase like 1 |
| chr6_-_8465656 | 0.87 |
ENSDART00000178887
|
si:dkey-217d24.6
|
si:dkey-217d24.6 |
| chr24_+_10039165 | 0.87 |
ENSDART00000144186
|
pou6f2
|
POU class 6 homeobox 2 |
| chr22_+_23430688 | 0.86 |
ENSDART00000160457
|
dennd1b
|
DENN/MADD domain containing 1B |
| chr24_-_2423791 | 0.85 |
ENSDART00000190402
|
rreb1a
|
ras responsive element binding protein 1a |
| chr12_-_25150239 | 0.85 |
ENSDART00000038415
ENSDART00000135368 |
rhoq
|
ras homolog family member Q |
| chr11_-_11961706 | 0.84 |
ENSDART00000115249
|
abcc10
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 10 |
| chr2_-_7185460 | 0.84 |
ENSDART00000092078
|
rc3h1b
|
ring finger and CCCH-type domains 1b |
| chr2_+_243778 | 0.84 |
ENSDART00000182262
|
CABZ01085887.1
|
|
| chr3_+_19685873 | 0.82 |
ENSDART00000006490
|
tlk2
|
tousled-like kinase 2 |
| chr21_+_3960583 | 0.82 |
ENSDART00000149788
|
setx
|
senataxin |
| chr8_+_3431671 | 0.82 |
ENSDART00000017850
|
ctu1
|
cytosolic thiouridylase subunit 1 homolog (S. pombe) |
| chr2_-_10632431 | 0.82 |
ENSDART00000122709
|
mtf2
|
metal response element binding transcription factor 2 |
| chr13_-_15799391 | 0.81 |
ENSDART00000124688
|
bag5
|
BCL2 associated athanogene 5 |
| chr16_-_25400257 | 0.80 |
ENSDART00000040756
|
zgc:136493
|
zgc:136493 |
| chr18_-_17145111 | 0.80 |
ENSDART00000090446
|
zc3h18
|
zinc finger CCCH-type containing 18 |
| chr11_-_25257595 | 0.79 |
ENSDART00000123567
|
snai1a
|
snail family zinc finger 1a |
| chr2_-_22530969 | 0.79 |
ENSDART00000159641
|
znf644a
|
zinc finger protein 644a |
| chr16_-_1709328 | 0.79 |
ENSDART00000168865
|
ascc3
|
activating signal cointegrator 1 complex subunit 3 |
| chr13_-_33317323 | 0.77 |
ENSDART00000110295
ENSDART00000144848 ENSDART00000136701 |
tmem234
|
transmembrane protein 234 |
| chr24_+_17069420 | 0.77 |
ENSDART00000014787
|
pip4k2aa
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha a |
| chr22_-_20924564 | 0.76 |
ENSDART00000100642
ENSDART00000032770 |
ell
|
elongation factor RNA polymerase II |
| chr7_+_39688208 | 0.76 |
ENSDART00000189682
|
tbc1d14
|
TBC1 domain family, member 14 |
| chr16_-_28594181 | 0.75 |
ENSDART00000059053
|
rpp38
|
ribonuclease P/MRP 38 subunit |
| chr16_-_22585289 | 0.74 |
ENSDART00000134239
ENSDART00000193959 ENSDART00000077998 |
si:dkey-238m4.3
cgna
|
si:dkey-238m4.3 cingulin a |
| chr16_+_11779534 | 0.74 |
ENSDART00000133497
|
si:dkey-250k15.4
|
si:dkey-250k15.4 |
| chr25_-_13408760 | 0.74 |
ENSDART00000154445
|
gins3
|
GINS complex subunit 3 |
| chr13_+_33368140 | 0.74 |
ENSDART00000033848
|
brf1a
|
BRF1, RNA polymerase III transcription initiation factor a |
| chr20_-_34750363 | 0.74 |
ENSDART00000152845
|
znf395b
|
zinc finger protein 395b |
| chr6_+_10338554 | 0.73 |
ENSDART00000186936
|
cobll1a
|
cordon-bleu WH2 repeat protein-like 1a |
| chr1_-_9858508 | 0.73 |
ENSDART00000147904
|
mad1l1
|
mitotic arrest deficient 1 like 1 |
| chr24_-_26518972 | 0.73 |
ENSDART00000097792
|
tnikb
|
TRAF2 and NCK interacting kinase b |
| chr22_-_3299100 | 0.73 |
ENSDART00000160305
|
si:zfos-943e10.1
|
si:zfos-943e10.1 |
| chr22_-_3299355 | 0.72 |
ENSDART00000190993
|
si:zfos-943e10.1
|
si:zfos-943e10.1 |
| chr4_-_26035770 | 0.72 |
ENSDART00000124514
|
usp44
|
ubiquitin specific peptidase 44 |
| chr14_-_24110062 | 0.72 |
ENSDART00000177062
|
cpeb4a
|
cytoplasmic polyadenylation element binding protein 4a |
| chr6_+_1724889 | 0.71 |
ENSDART00000157415
|
acvr2ab
|
activin A receptor type 2Ab |
| chr13_+_46941930 | 0.71 |
ENSDART00000056962
|
fbxo5
|
F-box protein 5 |
| chr10_+_41945890 | 0.71 |
ENSDART00000063013
ENSDART00000128313 |
tmem120b
|
transmembrane protein 120B |
| chr13_+_33368503 | 0.70 |
ENSDART00000139650
|
brf1a
|
BRF1, RNA polymerase III transcription initiation factor a |
| chr16_+_11779761 | 0.70 |
ENSDART00000140297
|
si:dkey-250k15.4
|
si:dkey-250k15.4 |
| chr8_-_12432604 | 0.70 |
ENSDART00000133350
ENSDART00000140699 ENSDART00000101174 |
traf1
|
TNF receptor-associated factor 1 |
| chr14_-_24110251 | 0.70 |
ENSDART00000079226
|
cpeb4a
|
cytoplasmic polyadenylation element binding protein 4a |
| chr22_-_34979139 | 0.70 |
ENSDART00000116455
ENSDART00000133537 |
arhgap19
|
Rho GTPase activating protein 19 |
| chr2_-_37280617 | 0.69 |
ENSDART00000190458
|
nadkb
|
NAD kinase b |
| chr20_-_28349144 | 0.69 |
ENSDART00000179690
ENSDART00000188059 |
ino80
|
INO80 complex subunit |
| chr7_+_22792895 | 0.69 |
ENSDART00000184407
|
rbm4.3
|
RNA binding motif protein 4.3 |
| chr15_-_30714130 | 0.69 |
ENSDART00000156914
ENSDART00000154714 |
msi2b
|
musashi RNA-binding protein 2b |
| chr5_-_10082244 | 0.69 |
ENSDART00000036421
|
chek2
|
checkpoint kinase 2 |
| chr5_+_13870340 | 0.68 |
ENSDART00000160690
|
hk2
|
hexokinase 2 |
| chr1_+_30723677 | 0.68 |
ENSDART00000177900
|
bora
|
bora, aurora kinase A activator |
| chr21_-_13225402 | 0.68 |
ENSDART00000080347
|
wdr34
|
WD repeat domain 34 |
| chr1_-_354115 | 0.68 |
ENSDART00000141590
ENSDART00000098627 |
pros1
|
protein S |
| chr13_+_15701596 | 0.67 |
ENSDART00000130832
|
trmt61a
|
tRNA methyltransferase 61A |
| chr7_+_53152108 | 0.67 |
ENSDART00000171350
|
cdh29
|
cadherin 29 |
| chr15_+_1766734 | 0.66 |
ENSDART00000168250
|
cul3b
|
cullin 3b |
| chr15_+_15266025 | 0.66 |
ENSDART00000112974
ENSDART00000184450 |
c2cd3
|
C2 calcium-dependent domain containing 3 |
| chr4_-_4570475 | 0.66 |
ENSDART00000184955
|
rassf3
|
Ras association (RalGDS/AF-6) domain family member 3 |
| chr16_-_42175617 | 0.66 |
ENSDART00000084715
|
alkbh8
|
alkB homolog 8, tRNA methyltransferase |
| chr22_+_2229964 | 0.66 |
ENSDART00000112582
|
znf1161
|
zinc finger protein 1161 |
| chr3_+_46315016 | 0.66 |
ENSDART00000157199
|
mkl2b
|
MKL/myocardin-like 2b |
| chr5_+_20035284 | 0.65 |
ENSDART00000191808
|
sgsm1a
|
small G protein signaling modulator 1a |
| chr2_+_51818039 | 0.65 |
ENSDART00000170353
|
acvr2bb
|
activin A receptor type 2Bb |
| chr22_+_6740039 | 0.65 |
ENSDART00000144122
|
CT583625.2
|
|
| chr9_+_19039608 | 0.65 |
ENSDART00000055866
|
chmp2ba
|
charged multivesicular body protein 2Ba |
| chr2_+_37424261 | 0.64 |
ENSDART00000132427
|
phc3
|
polyhomeotic homolog 3 (Drosophila) |
| chr4_-_77252368 | 0.64 |
ENSDART00000111941
|
zgc:174310
|
zgc:174310 |
| chr16_-_31284922 | 0.63 |
ENSDART00000142638
|
mroh1
|
maestro heat-like repeat family member 1 |
| chr11_-_27953135 | 0.63 |
ENSDART00000168338
|
ece1
|
endothelin converting enzyme 1 |
| chr19_+_14352332 | 0.62 |
ENSDART00000164386
|
arid1ab
|
AT rich interactive domain 1Ab (SWI-like) |
| chr16_-_4610255 | 0.62 |
ENSDART00000081852
ENSDART00000123253 ENSDART00000127554 ENSDART00000029485 |
arnt
|
aryl hydrocarbon receptor nuclear translocator |
| chr13_+_8840772 | 0.62 |
ENSDART00000059321
|
epcam
|
epithelial cell adhesion molecule |
| chr15_-_28569786 | 0.62 |
ENSDART00000127845
|
ssh2a
|
slingshot protein phosphatase 2a |
| chr16_-_39267185 | 0.61 |
ENSDART00000058550
ENSDART00000133642 |
gpd1l
|
glycerol-3-phosphate dehydrogenase 1 like |
| chr16_-_40459104 | 0.61 |
ENSDART00000032389
|
plekhf2
|
pleckstrin homology domain containing, family F (with FYVE domain) member 2 |
| chr10_-_10018120 | 0.60 |
ENSDART00000132375
|
strbp
|
spermatid perinuclear RNA binding protein |
| chr7_+_41887429 | 0.60 |
ENSDART00000115090
|
gpt2
|
glutamic pyruvate transaminase (alanine aminotransferase) 2 |
| chr3_-_31619463 | 0.60 |
ENSDART00000124559
|
moto
|
minamoto |
| chr7_-_64770456 | 0.60 |
ENSDART00000192618
|
zdhhc21
|
zinc finger, DHHC-type containing 21 |
| chr10_+_44373349 | 0.59 |
ENSDART00000172191
|
snrnp35
|
small nuclear ribonucleoprotein 35 (U11/U12) |
| chr18_+_20468157 | 0.59 |
ENSDART00000100665
ENSDART00000147867 ENSDART00000060302 ENSDART00000180370 |
ddb2
|
damage-specific DNA binding protein 2 |
| chr16_+_3185541 | 0.59 |
ENSDART00000024088
|
wdr21
|
WD repeat domain 21 |
| chr3_-_11878490 | 0.59 |
ENSDART00000129961
|
coro7
|
coronin 7 |
| chr19_+_24896409 | 0.59 |
ENSDART00000049840
|
eya3
|
EYA transcriptional coactivator and phosphatase 3 |
| chr8_-_49304602 | 0.58 |
ENSDART00000147020
|
prickle3
|
prickle homolog 3 |
| chr22_-_24757785 | 0.58 |
ENSDART00000078225
|
vtg5
|
vitellogenin 5 |
| chr18_+_20225961 | 0.58 |
ENSDART00000045679
|
tle3a
|
transducin-like enhancer of split 3a |
| chr21_+_76739 | 0.57 |
ENSDART00000174654
|
ARSB
|
arylsulfatase B |
| chr16_+_17763848 | 0.56 |
ENSDART00000149408
ENSDART00000148878 |
them4
|
thioesterase superfamily member 4 |
| chr5_-_54481692 | 0.56 |
ENSDART00000165719
|
fbxw5
|
F-box and WD repeat domain containing 5 |
| chr11_+_34522554 | 0.56 |
ENSDART00000109833
|
zmat3
|
zinc finger, matrin-type 3 |
| chr1_-_51261420 | 0.56 |
ENSDART00000168685
|
kif16ba
|
kinesin family member 16Ba |
| chr19_+_4051695 | 0.55 |
ENSDART00000166368
|
btr24
|
bloodthirsty-related gene family, member 24 |
| chr15_-_17169935 | 0.55 |
ENSDART00000110111
|
cul5a
|
cullin 5a |
| chr9_-_46072805 | 0.55 |
ENSDART00000169682
|
hdac4
|
histone deacetylase 4 |
| chr2_+_31437547 | 0.55 |
ENSDART00000141170
|
stam
|
signal transducing adaptor molecule (SH3 domain and ITAM motif) 1 |
| chr7_+_71586485 | 0.55 |
ENSDART00000165582
|
smchd1
|
structural maintenance of chromosomes flexible hinge domain containing 1 |
| chr12_+_33361948 | 0.54 |
ENSDART00000124982
|
fasn
|
fatty acid synthase |
| chr10_-_41156348 | 0.54 |
ENSDART00000058622
|
aak1b
|
AP2 associated kinase 1b |
| chr20_-_9194257 | 0.54 |
ENSDART00000133012
|
ylpm1
|
YLP motif containing 1 |
| chr23_-_24952264 | 0.54 |
ENSDART00000192085
|
zbtb48
|
zinc finger and BTB domain containing 48 |
| chr9_-_17783574 | 0.54 |
ENSDART00000146706
|
vwa8
|
von Willebrand factor A domain containing 8 |
| chr24_-_31332087 | 0.53 |
ENSDART00000161179
|
abcd3a
|
ATP-binding cassette, sub-family D (ALD), member 3a |
| chr12_+_33320884 | 0.52 |
ENSDART00000188988
|
csnk1db
|
casein kinase 1, delta b |
| chr19_+_4066449 | 0.52 |
ENSDART00000162461
|
btr26
|
bloodthirsty-related gene family, member 26 |
| chr12_+_34770531 | 0.52 |
ENSDART00000153320
|
slc38a10
|
solute carrier family 38, member 10 |
| chr18_+_47301086 | 0.52 |
ENSDART00000009775
|
rbm7
|
RNA binding motif protein 7 |
| chr3_-_31875138 | 0.52 |
ENSDART00000155298
|
limd2
|
LIM domain containing 2 |
| chr24_+_37080771 | 0.52 |
ENSDART00000159942
|
kcnc3b
|
potassium voltage-gated channel, Shaw-related subfamily, member 3b |
| chr2_+_2967255 | 0.51 |
ENSDART00000167649
ENSDART00000166449 |
pik3r3a
|
phosphoinositide-3-kinase, regulatory subunit 3a (gamma) |
| chr16_-_12097394 | 0.51 |
ENSDART00000103944
|
pex5
|
peroxisomal biogenesis factor 5 |
| chr18_+_17600570 | 0.51 |
ENSDART00000175258
ENSDART00000151850 ENSDART00000151934 |
herpud1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr6_-_31739709 | 0.51 |
ENSDART00000087964
|
cachd1
|
cache domain containing 1 |
| chr16_-_12097558 | 0.51 |
ENSDART00000123142
|
pex5
|
peroxisomal biogenesis factor 5 |
| chr21_-_37951819 | 0.51 |
ENSDART00000139549
|
si:dkey-38k9.5
|
si:dkey-38k9.5 |
| chr11_-_34522249 | 0.50 |
ENSDART00000158616
|
pik3ca
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha |
| chr21_-_18086114 | 0.50 |
ENSDART00000143987
|
vav2
|
vav 2 guanine nucleotide exchange factor |
| chr2_+_38924975 | 0.50 |
ENSDART00000109219
|
rem2
|
RAS (RAD and GEM)-like GTP binding 2 |
| chr20_-_53963515 | 0.49 |
ENSDART00000110252
|
ppp2r5cb
|
protein phosphatase 2, regulatory subunit B', gamma b |
| chr9_-_27748868 | 0.49 |
ENSDART00000190306
|
tbccd1
|
TBCC domain containing 1 |
| chr13_-_42536642 | 0.48 |
ENSDART00000134533
|
btaf1
|
BTAF1 RNA polymerase II, B-TFIID transcription factor-associated |
| chr20_+_812012 | 0.48 |
ENSDART00000179299
ENSDART00000137135 |
mei4
|
meiosis-specific, MEI4 homolog (S. cerevisiae) |
| chr19_-_10656667 | 0.48 |
ENSDART00000081379
ENSDART00000151456 ENSDART00000143271 ENSDART00000182126 |
olah
|
oleoyl-ACP hydrolase |
| chr22_-_8006342 | 0.47 |
ENSDART00000162028
|
sc:d217
|
sc:d217 |
| chr2_+_36121373 | 0.47 |
ENSDART00000187002
|
CT867973.2
|
|
| chr5_-_55914268 | 0.47 |
ENSDART00000014049
|
wdr36
|
WD repeat domain 36 |
| chr17_+_52612866 | 0.47 |
ENSDART00000182828
ENSDART00000191156 ENSDART00000188814 ENSDART00000109891 |
angel1
|
angel homolog 1 (Drosophila) |
| chr4_-_72287764 | 0.47 |
ENSDART00000125452
ENSDART00000189437 |
si:cabz01071911.3
|
si:cabz01071911.3 |
| chr16_-_33105677 | 0.46 |
ENSDART00000145055
|
pnrc2
|
proline-rich nuclear receptor coactivator 2 |
| chr21_-_27221535 | 0.46 |
ENSDART00000138266
|
mark2a
|
MAP/microtubule affinity-regulating kinase 2a |
| chr7_+_5956937 | 0.46 |
ENSDART00000170763
|
hist1h2a4
|
histone cluster 1 H2A family member 4 |
| chr20_+_53368611 | 0.45 |
ENSDART00000060432
|
cdc40
|
cell division cycle 40 homolog (S. cerevisiae) |
| chr5_+_32009080 | 0.45 |
ENSDART00000186885
|
scai
|
suppressor of cancer cell invasion |
| chr3_+_16668609 | 0.45 |
ENSDART00000185646
|
zgc:55558
|
zgc:55558 |
| chr17_-_35279422 | 0.45 |
ENSDART00000181587
|
adam17a
|
ADAM metallopeptidase domain 17a |
| chr1_-_28089557 | 0.44 |
ENSDART00000161024
ENSDART00000167875 |
snapc3
|
small nuclear RNA activating complex, polypeptide 3 |
| chr25_-_10615504 | 0.44 |
ENSDART00000156797
|
ppp6r3
|
protein phosphatase 6, regulatory subunit 3 |
| chr1_+_41690402 | 0.44 |
ENSDART00000177298
|
fbxo41
|
F-box protein 41 |
| chr16_-_8132742 | 0.43 |
ENSDART00000104323
|
snrka
|
SNF related kinase a |
| chr7_+_21331688 | 0.43 |
ENSDART00000128014
|
dnah2
|
dynein, axonemal, heavy chain 2 |
| chr2_-_22286828 | 0.43 |
ENSDART00000168653
|
fam110b
|
family with sequence similarity 110, member B |
| chr2_-_10877228 | 0.43 |
ENSDART00000138718
ENSDART00000034246 |
cdc7
|
cell division cycle 7 homolog (S. cerevisiae) |
| chr19_-_4851411 | 0.43 |
ENSDART00000110398
|
fbxl20
|
F-box and leucine-rich repeat protein 20 |
| chr2_+_33052360 | 0.43 |
ENSDART00000134651
|
rnf220a
|
ring finger protein 220a |
| chr20_+_51833030 | 0.43 |
ENSDART00000074330
|
taf1a
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, A |
| chr3_-_26805455 | 0.43 |
ENSDART00000180648
|
pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
Network of associatons between targets according to the STRING database.
First level regulatory network of nkx2.2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.2 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.5 | 1.4 | GO:0005991 | trehalose metabolic process(GO:0005991) |
| 0.4 | 1.6 | GO:0015878 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.4 | 2.0 | GO:0010719 | photoreceptor cell morphogenesis(GO:0008594) negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.4 | 2.6 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.4 | 1.4 | GO:0051352 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.3 | 1.5 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.3 | 2.5 | GO:0043696 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.3 | 1.1 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.2 | 0.7 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.2 | 0.9 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.2 | 1.2 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.2 | 1.8 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.2 | 1.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.2 | 0.5 | GO:0090299 | regulation of neural crest formation(GO:0090299) |
| 0.2 | 0.9 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.2 | 0.9 | GO:0030576 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) protein localization to nucleoplasm(GO:1990173) |
| 0.2 | 0.5 | GO:0021698 | cerebellar Purkinje cell layer structural organization(GO:0021693) cerebellar cortex structural organization(GO:0021698) |
| 0.2 | 0.5 | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) |
| 0.2 | 0.6 | GO:0006116 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.1 | 0.9 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 1.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 0.7 | GO:0042766 | nucleosome mobilization(GO:0042766) |
| 0.1 | 1.9 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.1 | 1.0 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.8 | GO:0032447 | tRNA wobble position uridine thiolation(GO:0002143) protein urmylation(GO:0032447) |
| 0.1 | 0.9 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.1 | 0.4 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.1 | 0.5 | GO:0051645 | Golgi localization(GO:0051645) maintenance of centrosome location(GO:0051661) |
| 0.1 | 0.4 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.1 | 0.8 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 0.3 | GO:0019557 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.1 | 1.1 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.1 | 0.7 | GO:1902315 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.1 | 0.3 | GO:0035971 | peptidyl-histidine dephosphorylation(GO:0035971) |
| 0.1 | 1.0 | GO:0022615 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.1 | 0.4 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.1 | 1.2 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 2.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 0.8 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.1 | 0.6 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.1 | 0.5 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.1 | 0.6 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.1 | 1.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.2 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.1 | 0.3 | GO:2000144 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.1 | 0.4 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.1 | 0.2 | GO:0048340 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) intermediate mesoderm development(GO:0048389) |
| 0.1 | 2.4 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.1 | 0.7 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.1 | 1.4 | GO:0007257 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.1 | 1.4 | GO:0070897 | DNA-templated transcriptional preinitiation complex assembly(GO:0070897) |
| 0.1 | 0.9 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) |
| 0.1 | 0.5 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 | 0.2 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.1 | 0.5 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.1 | 0.4 | GO:0032205 | negative regulation of telomere maintenance(GO:0032205) |
| 0.1 | 0.6 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.1 | 0.3 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.1 | 0.4 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.2 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.1 | 0.3 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 0.5 | GO:0032204 | regulation of telomere maintenance(GO:0032204) |
| 0.1 | 0.8 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.1 | 0.9 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.3 | GO:0090134 | establishment or maintenance of cytoskeleton polarity(GO:0030952) mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.0 | 1.1 | GO:0005979 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.0 | 0.8 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.0 | 0.7 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.0 | 0.2 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.7 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.2 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 0.0 | 0.7 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.3 | GO:0009146 | dGTP catabolic process(GO:0006203) purine nucleoside triphosphate catabolic process(GO:0009146) purine deoxyribonucleoside triphosphate catabolic process(GO:0009217) dGTP metabolic process(GO:0046070) |
| 0.0 | 0.4 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.3 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.5 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.7 | GO:0010824 | regulation of centrosome duplication(GO:0010824) |
| 0.0 | 0.2 | GO:0015990 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) electron transport coupled proton transport(GO:0015990) |
| 0.0 | 0.5 | GO:0048696 | collateral sprouting in absence of injury(GO:0048669) regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.0 | 0.4 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.0 | 0.4 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.2 | GO:0030823 | regulation of cGMP metabolic process(GO:0030823) positive regulation of cGMP metabolic process(GO:0030825) regulation of cGMP biosynthetic process(GO:0030826) positive regulation of cGMP biosynthetic process(GO:0030828) regulation of guanylate cyclase activity(GO:0031282) positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.1 | GO:0042543 | protein N-linked glycosylation via arginine(GO:0042543) |
| 0.0 | 0.7 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.4 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.1 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.2 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 1.0 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.0 | 0.4 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.2 | GO:0090200 | regulation of release of cytochrome c from mitochondria(GO:0090199) positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.3 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.0 | 0.4 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 1.4 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.0 | 0.1 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 0.1 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.2 | GO:1901339 | regulation of store-operated calcium channel activity(GO:1901339) |
| 0.0 | 0.6 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) late endosome to vacuole transport(GO:0045324) |
| 0.0 | 0.7 | GO:0044773 | mitotic DNA damage checkpoint(GO:0044773) |
| 0.0 | 0.2 | GO:0015809 | arginine transport(GO:0015809) |
| 0.0 | 0.2 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.4 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.6 | GO:0009411 | response to UV(GO:0009411) |
| 0.0 | 0.5 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 0.4 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.2 | GO:0003188 | heart valve formation(GO:0003188) atrioventricular valve formation(GO:0003190) |
| 0.0 | 0.1 | GO:0009120 | deoxyribonucleoside metabolic process(GO:0009120) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.0 | 0.7 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.8 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 1.1 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.4 | GO:2000404 | regulation of T cell migration(GO:2000404) |
| 0.0 | 0.5 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.1 | GO:1904666 | regulation of ubiquitin protein ligase activity(GO:1904666) |
| 0.0 | 0.1 | GO:0043649 | dicarboxylic acid catabolic process(GO:0043649) |
| 0.0 | 0.6 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.2 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.1 | GO:0060465 | pharynx development(GO:0060465) |
| 0.0 | 0.6 | GO:0048920 | posterior lateral line neuromast primordium migration(GO:0048920) |
| 0.0 | 0.9 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.5 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.1 | GO:0006478 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 0.5 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.6 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.0 | 1.2 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.2 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.4 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.4 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.8 | GO:0060968 | regulation of gene silencing(GO:0060968) |
| 0.0 | 0.6 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.0 | 0.5 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.0 | 0.5 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.0 | 0.6 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 1.0 | GO:0050863 | regulation of T cell activation(GO:0050863) regulation of leukocyte cell-cell adhesion(GO:1903037) |
| 0.0 | 0.1 | GO:0008584 | male gonad development(GO:0008584) |
| 0.0 | 0.5 | GO:0042752 | regulation of circadian rhythm(GO:0042752) |
| 0.0 | 0.1 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.4 | GO:0006360 | transcription from RNA polymerase I promoter(GO:0006360) |
| 0.0 | 0.4 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.3 | GO:0060173 | limb development(GO:0060173) |
| 0.0 | 0.1 | GO:1902047 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.3 | GO:0007520 | myoblast fusion(GO:0007520) regulation of neutrophil migration(GO:1902622) |
| 0.0 | 0.3 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.0 | 0.2 | GO:0086010 | membrane depolarization(GO:0051899) membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.1 | GO:1901099 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.6 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.5 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.8 | GO:0030681 | fibrillar center(GO:0001650) multimeric ribonuclease P complex(GO:0030681) |
| 0.2 | 0.9 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.2 | 0.9 | GO:1990923 | PET complex(GO:1990923) |
| 0.2 | 0.6 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.2 | 1.4 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.2 | 2.6 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.7 | GO:0000811 | GINS complex(GO:0000811) |
| 0.1 | 0.4 | GO:0019185 | snRNA-activating protein complex(GO:0019185) |
| 0.1 | 0.4 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.1 | 0.7 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.1 | 0.9 | GO:0016234 | inclusion body(GO:0016234) |
| 0.1 | 0.7 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.1 | 0.7 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 0.6 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 1.4 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 0.5 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 0.6 | GO:0035060 | brahma complex(GO:0035060) |
| 0.1 | 1.4 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.2 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.1 | 0.5 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 0.4 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.1 | 1.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.8 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 1.2 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 1.5 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.5 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.9 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.6 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 4.6 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.6 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.3 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 1.2 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 1.5 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.3 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.5 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.6 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.5 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.7 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.5 | GO:0036452 | ESCRT complex(GO:0036452) |
| 0.0 | 2.3 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.5 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.4 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.4 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.7 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 2.0 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 2.1 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.8 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 1.3 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.4 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.4 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.1 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.1 | GO:0034451 | acrosomal vesicle(GO:0001669) centriolar satellite(GO:0034451) |
| 0.0 | 0.7 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.8 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 1.6 | GO:0031461 | cullin-RING ubiquitin ligase complex(GO:0031461) |
| 0.0 | 0.6 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.5 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0004555 | alpha,alpha-trehalase activity(GO:0004555) trehalase activity(GO:0015927) |
| 0.4 | 1.8 | GO:0033204 | ribonuclease P RNA binding(GO:0033204) |
| 0.4 | 1.6 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.3 | 0.8 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.3 | 0.8 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.2 | 1.9 | GO:0001016 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 3 promoter sequence-specific DNA binding(GO:0001006) RNA polymerase III regulatory region DNA binding(GO:0001016) RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.2 | 0.7 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.2 | 1.1 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.2 | 0.8 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.2 | 0.8 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.2 | 2.6 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.2 | 1.4 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.2 | 0.6 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.1 | 0.7 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.1 | 1.0 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.1 | 0.8 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 1.1 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.1 | 2.0 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 0.9 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.1 | 0.3 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 0.7 | GO:0005536 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 0.7 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 0.9 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 0.1 | 0.4 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 2.2 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.1 | 1.1 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 1.5 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.1 | 0.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 1.4 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.1 | 0.6 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.1 | 0.8 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 1.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.1 | 0.3 | GO:0101006 | protein histidine phosphatase activity(GO:0101006) |
| 0.1 | 0.2 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.1 | 0.2 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.1 | 1.1 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 0.3 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.1 | 0.4 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.1 | 0.3 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.1 | 0.3 | GO:0016793 | dGTPase activity(GO:0008832) triphosphoric monoester hydrolase activity(GO:0016793) |
| 0.0 | 1.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.6 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.4 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.5 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.5 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 3.0 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 0.5 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.2 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
| 0.0 | 0.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.5 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.2 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.0 | 0.4 | GO:0052794 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.9 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.0 | 2.4 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.8 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.2 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.1 | GO:0047325 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) |
| 0.0 | 0.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.1 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 1.0 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 1.1 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.5 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.5 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.2 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.0 | 0.5 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.8 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.9 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 1.4 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 0.6 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 0.8 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.8 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.6 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.5 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.5 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 1.0 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.0 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.1 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.5 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 1.1 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.3 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.6 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 2.7 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 2.6 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.4 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 1.4 | GO:1901981 | phosphatidylinositol phosphate binding(GO:1901981) |
| 0.0 | 0.6 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 3.0 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.3 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.2 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.4 | GO:0019903 | protein phosphatase binding(GO:0019903) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 0.5 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 0.8 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 1.7 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 1.0 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.1 | 2.2 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 1.6 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.9 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.7 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.4 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.5 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.5 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.5 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.2 | 0.7 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.4 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.1 | 0.6 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 1.0 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 0.6 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 0.6 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 0.7 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 0.7 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.9 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.6 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.5 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.4 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.6 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 1.3 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.7 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.8 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.6 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 1.1 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.4 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.5 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.5 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.4 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 1.3 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.8 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.5 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.4 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 0.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.4 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.2 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |