Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
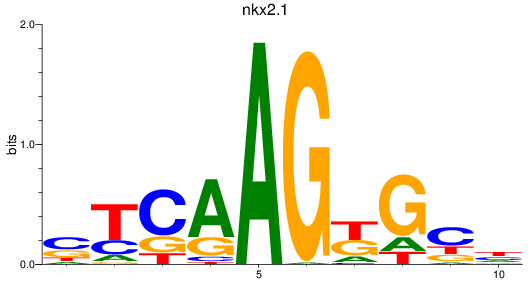
Results for nkx2.1
Z-value: 1.10
Transcription factors associated with nkx2.1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nkx2.1
|
ENSDARG00000019835 | NK2 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nkx2.1 | dr11_v1_chr17_+_38262408_38262408 | -0.95 | 9.6e-05 | Click! |
Activity profile of nkx2.1 motif
Sorted Z-values of nkx2.1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_24597001 | 1.72 |
ENSDART00000191834
|
rlf
|
rearranged L-myc fusion |
| chr7_+_34602991 | 1.56 |
ENSDART00000073447
|
fhod1
|
formin homology 2 domain containing 1 |
| chr6_+_38880166 | 1.51 |
ENSDART00000019939
ENSDART00000144286 |
bin2b
|
bridging integrator 2b |
| chr21_+_34132747 | 1.48 |
ENSDART00000148115
|
trpc5b
|
transient receptor potential cation channel, subfamily C, member 5b |
| chr8_-_44299247 | 1.48 |
ENSDART00000144497
|
piwil1
|
piwi-like RNA-mediated gene silencing 1 |
| chr6_+_38879961 | 1.40 |
ENSDART00000184798
|
bin2b
|
bridging integrator 2b |
| chr4_-_14531687 | 1.37 |
ENSDART00000182093
ENSDART00000159447 |
plxnb2a
|
plexin b2a |
| chr3_-_23461954 | 1.35 |
ENSDART00000040065
|
casc3
|
cancer susceptibility candidate 3 |
| chr22_+_8753092 | 1.31 |
ENSDART00000140720
|
si:dkey-182g1.2
|
si:dkey-182g1.2 |
| chr14_-_33478963 | 1.30 |
ENSDART00000132813
|
lamp2
|
lysosomal-associated membrane protein 2 |
| chr12_+_30788912 | 1.30 |
ENSDART00000160422
|
aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr20_+_9128829 | 1.29 |
ENSDART00000064144
ENSDART00000137450 |
bpnt1
|
bisphosphate nucleotidase 1 |
| chr5_-_54712159 | 1.26 |
ENSDART00000149207
|
ccnb1
|
cyclin B1 |
| chr18_+_44631789 | 1.24 |
ENSDART00000144271
|
bicra
|
BRD4 interacting chromatin remodeling complex associated protein |
| chr11_-_36040549 | 1.10 |
ENSDART00000112684
|
setmar
|
SET domain and mariner transposase fusion gene |
| chr3_-_21166597 | 1.10 |
ENSDART00000175941
|
taok2a
|
TAO kinase 2a |
| chr7_-_24875421 | 1.08 |
ENSDART00000173920
|
adad2
|
adenosine deaminase domain containing 2 |
| chr15_-_29012493 | 1.08 |
ENSDART00000060018
|
dharma
|
dharma |
| chr13_+_10023256 | 1.05 |
ENSDART00000110035
|
srbd1
|
S1 RNA binding domain 1 |
| chr3_+_39600562 | 1.02 |
ENSDART00000134309
ENSDART00000007170 |
prpsap2
|
phosphoribosyl pyrophosphate synthetase-associated protein 2 |
| chr7_-_59123066 | 0.98 |
ENSDART00000175438
|
dennd4c
|
DENN/MADD domain containing 4C |
| chr17_-_12249990 | 0.96 |
ENSDART00000177889
ENSDART00000155545 |
ahctf1
|
AT hook containing transcription factor 1 |
| chr7_-_64770456 | 0.94 |
ENSDART00000192618
|
zdhhc21
|
zinc finger, DHHC-type containing 21 |
| chr3_-_30888415 | 0.94 |
ENSDART00000124458
|
kmt5c
|
lysine methyltransferase 5C |
| chr1_+_16621345 | 0.93 |
ENSDART00000149026
|
pcm1
|
pericentriolar material 1 |
| chr8_-_4100365 | 0.92 |
ENSDART00000142846
|
cux2b
|
cut-like homeobox 2b |
| chr1_+_27977297 | 0.91 |
ENSDART00000180692
ENSDART00000166819 |
sugt1
|
SGT1 homolog, MIS12 kinetochore complex assembly cochaperone |
| chr23_-_33558161 | 0.91 |
ENSDART00000018301
|
itga5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide) |
| chr4_-_8152746 | 0.83 |
ENSDART00000012928
ENSDART00000177482 |
wnk1b
|
WNK lysine deficient protein kinase 1b |
| chr3_+_17933132 | 0.80 |
ENSDART00000104299
ENSDART00000162144 ENSDART00000162242 ENSDART00000166289 ENSDART00000171101 ENSDART00000164853 |
cnp
|
2',3'-cyclic nucleotide 3' phosphodiesterase |
| chr6_-_18531760 | 0.77 |
ENSDART00000167167
|
utp6
|
UTP6, small subunit (SSU) processome component, homolog (yeast) |
| chr3_+_22335030 | 0.77 |
ENSDART00000055676
|
zgc:103564
|
zgc:103564 |
| chr25_-_17590971 | 0.77 |
ENSDART00000189942
|
mmp15a
|
matrix metallopeptidase 15a |
| chr11_+_25596038 | 0.74 |
ENSDART00000140856
|
ccdc120
|
coiled-coil domain containing 120 |
| chr7_-_23777445 | 0.72 |
ENSDART00000173527
|
si:ch211-200p22.4
|
si:ch211-200p22.4 |
| chr7_-_48733662 | 0.71 |
ENSDART00000191675
|
traf6
|
TNF receptor-associated factor 6 |
| chr17_+_33999630 | 0.69 |
ENSDART00000167085
ENSDART00000155030 ENSDART00000168522 ENSDART00000191799 ENSDART00000189684 ENSDART00000153942 ENSDART00000187272 ENSDART00000127692 |
gphna
|
gephyrin a |
| chr15_-_434503 | 0.67 |
ENSDART00000122286
|
CABZ01056629.1
|
|
| chr2_+_314249 | 0.65 |
ENSDART00000082086
|
zgc:113452
|
zgc:113452 |
| chr21_+_45502621 | 0.65 |
ENSDART00000166719
|
si:dkey-223p19.2
|
si:dkey-223p19.2 |
| chr25_-_20691075 | 0.65 |
ENSDART00000067373
|
edc3
|
enhancer of mRNA decapping 3 homolog (S. cerevisiae) |
| chr15_-_23482088 | 0.64 |
ENSDART00000185823
ENSDART00000185523 |
nlrx1
|
NLR family member X1 |
| chr22_-_5724085 | 0.64 |
ENSDART00000110526
ENSDART00000140905 |
pgbd4
|
piggyBac transposable element derived 4 |
| chr18_-_46020138 | 0.63 |
ENSDART00000027772
|
pcyt1ab
|
phosphate cytidylyltransferase 1, choline, alpha b |
| chr21_-_2348838 | 0.62 |
ENSDART00000160337
|
si:ch73-299h12.8
|
si:ch73-299h12.8 |
| chr13_-_10727550 | 0.61 |
ENSDART00000190925
|
ppm1ba
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Ba |
| chr2_-_37401600 | 0.61 |
ENSDART00000015723
|
prkci
|
protein kinase C, iota |
| chr22_+_39096911 | 0.58 |
ENSDART00000157127
ENSDART00000153841 |
lmcd1
|
LIM and cysteine-rich domains 1 |
| chr20_-_38827623 | 0.57 |
ENSDART00000153310
|
cad
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr7_+_1442059 | 0.54 |
ENSDART00000173391
|
si:cabz01090193.1
|
si:cabz01090193.1 |
| chr7_+_48555626 | 0.54 |
ENSDART00000125483
ENSDART00000083514 |
kcnq1
|
potassium voltage-gated channel, KQT-like subfamily, member 1 |
| chr3_+_17933553 | 0.53 |
ENSDART00000167731
ENSDART00000165644 |
cnp
|
2',3'-cyclic nucleotide 3' phosphodiesterase |
| chr5_-_35456269 | 0.53 |
ENSDART00000051312
|
ttc33
|
tetratricopeptide repeat domain 33 |
| chr7_+_25825667 | 0.48 |
ENSDART00000149835
|
mtm1
|
myotubularin 1 |
| chr1_-_53756851 | 0.48 |
ENSDART00000122445
|
akt3b
|
v-akt murine thymoma viral oncogene homolog 3b |
| chr2_-_8609653 | 0.47 |
ENSDART00000193354
ENSDART00000189489 ENSDART00000186144 |
si:ch211-71m22.1
|
si:ch211-71m22.1 |
| chr7_+_48675347 | 0.47 |
ENSDART00000157917
ENSDART00000185580 |
trpm5
|
transient receptor potential cation channel, subfamily M, member 5 |
| chr24_+_39576071 | 0.46 |
ENSDART00000147137
|
si:dkey-161j23.6
|
si:dkey-161j23.6 |
| chr7_+_24390939 | 0.45 |
ENSDART00000087494
ENSDART00000125463 |
haus3
|
HAUS augmin-like complex, subunit 3 |
| chr21_+_34088377 | 0.45 |
ENSDART00000170070
|
mtmr1b
|
myotubularin related protein 1b |
| chr16_-_29397395 | 0.43 |
ENSDART00000130757
|
tlr18
|
toll-like receptor 18 |
| chr4_-_9054947 | 0.43 |
ENSDART00000109764
|
si:dkey-48p11.3
|
si:dkey-48p11.3 |
| chr13_+_46927350 | 0.43 |
ENSDART00000165041
ENSDART00000167931 |
mtrf1l
|
mitochondrial translational release factor 1-like |
| chr7_+_48555400 | 0.43 |
ENSDART00000174474
|
kcnq1
|
potassium voltage-gated channel, KQT-like subfamily, member 1 |
| chr11_-_24063196 | 0.43 |
ENSDART00000036513
|
trib3
|
tribbles pseudokinase 3 |
| chr14_+_10656975 | 0.42 |
ENSDART00000127594
ENSDART00000125865 |
atrx
|
alpha thalassemia/mental retardation syndrome X-linked homolog (human) |
| chr8_-_20243389 | 0.42 |
ENSDART00000184904
|
acer1
|
alkaline ceramidase 1 |
| chr2_+_26240339 | 0.42 |
ENSDART00000191006
|
palm1b
|
paralemmin 1b |
| chr20_+_46255057 | 0.40 |
ENSDART00000100536
|
taar14i
|
trace amine associated receptor 14i |
| chr2_+_19195841 | 0.39 |
ENSDART00000163137
ENSDART00000161095 |
elovl1a
|
ELOVL fatty acid elongase 1a |
| chr2_-_11119303 | 0.35 |
ENSDART00000135450
ENSDART00000131836 |
cryz
|
crystallin, zeta (quinone reductase) |
| chr12_-_19862912 | 0.32 |
ENSDART00000145788
|
shisa9a
|
shisa family member 9a |
| chr6_+_18531932 | 0.30 |
ENSDART00000165271
|
suz12b
|
SUZ12 polycomb repressive complex 2b subunit |
| chr14_-_10321851 | 0.29 |
ENSDART00000143382
|
il1rapl2
|
interleukin 1 receptor accessory protein-like 2 |
| chr2_+_26237322 | 0.28 |
ENSDART00000030520
|
palm1b
|
paralemmin 1b |
| chr18_+_45504362 | 0.28 |
ENSDART00000140089
|
cngb1a
|
cyclic nucleotide gated channel beta 1a |
| chr12_+_34953038 | 0.27 |
ENSDART00000187022
ENSDART00000123988 ENSDART00000027034 |
qki2
|
QKI, KH domain containing, RNA binding 2 |
| chr3_-_4455951 | 0.24 |
ENSDART00000193908
ENSDART00000074077 |
trim35-3
|
tripartite motif containing 35-3 |
| chr22_+_28818291 | 0.23 |
ENSDART00000136032
|
tp53bp2b
|
tumor protein p53 binding protein, 2b |
| chr1_+_8304904 | 0.22 |
ENSDART00000168631
|
cacna1hb
|
calcium channel, voltage-dependent, T type, alpha 1H subunit b |
| chr3_-_7464250 | 0.22 |
ENSDART00000159873
|
znf1001
|
zinc finger protein 1001 |
| chr4_-_50926767 | 0.21 |
ENSDART00000183430
|
si:ch211-208f21.3
|
si:ch211-208f21.3 |
| chr19_-_6983002 | 0.21 |
ENSDART00000104891
|
znf384l
|
zinc finger protein 384 like |
| chr18_-_22701800 | 0.20 |
ENSDART00000135098
|
si:ch73-113g13.3
|
si:ch73-113g13.3 |
| chr4_-_2052687 | 0.18 |
ENSDART00000138291
ENSDART00000150844 |
cpsf6
|
cleavage and polyadenylation specific factor 6 |
| chr4_-_20314749 | 0.14 |
ENSDART00000066894
ENSDART00000188123 |
dcp1b
|
decapping mRNA 1B |
| chr1_-_23294753 | 0.13 |
ENSDART00000013263
|
ugdh
|
UDP-glucose 6-dehydrogenase |
| chr13_+_771403 | 0.11 |
ENSDART00000093166
|
nrxn1b
|
neurexin 1b |
| chr15_-_31357634 | 0.11 |
ENSDART00000127485
|
or111-2
|
odorant receptor, family D, subfamily 111, member 2 |
| chr1_+_55662491 | 0.11 |
ENSDART00000152386
|
adgre8
|
adhesion G protein-coupled receptor E8 |
| chr1_-_43862638 | 0.11 |
ENSDART00000145044
|
tacr3a
|
tachykinin receptor 3a |
| chr18_-_39321484 | 0.11 |
ENSDART00000077694
|
leo1
|
LEO1 homolog, Paf1/RNA polymerase II complex component |
| chr11_+_12879635 | 0.10 |
ENSDART00000182515
ENSDART00000081296 |
si:dkey-11m19.5
|
si:dkey-11m19.5 |
| chr8_-_36469117 | 0.10 |
ENSDART00000111240
|
mhc2dab
|
major histocompatibility complex class II DAB gene |
| chr11_+_37909654 | 0.09 |
ENSDART00000172211
|
si:ch211-112f3.4
|
si:ch211-112f3.4 |
| chr3_+_32118670 | 0.05 |
ENSDART00000055287
ENSDART00000111688 |
zgc:109934
|
zgc:109934 |
| chr3_+_17846890 | 0.03 |
ENSDART00000193384
|
znf385c
|
zinc finger protein 385C |
| chr7_-_30639385 | 0.03 |
ENSDART00000173618
|
myo1ea
|
myosin IE, a |
| chr11_-_1956204 | 0.03 |
ENSDART00000185541
|
nr1d4b
|
nuclear receptor subfamily 1, group D, member 4b |
| chr2_-_36494308 | 0.02 |
ENSDART00000110378
|
BX901889.2
|
|
| chr11_-_8271374 | 0.01 |
ENSDART00000168253
|
pimr202
|
Pim proto-oncogene, serine/threonine kinase, related 202 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nkx2.1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.9 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.3 | 0.9 | GO:0034770 | histone H4-K20 methylation(GO:0034770) histone H4-K20 trimethylation(GO:0034773) |
| 0.3 | 1.0 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.2 | 1.5 | GO:0035093 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.2 | 0.7 | GO:0018315 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.2 | 1.1 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 0.2 | 1.0 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.2 | 0.6 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) |
| 0.2 | 0.9 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.2 | 0.5 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.2 | 0.9 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.2 | 0.6 | GO:0045217 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) cell-cell junction maintenance(GO:0045217) |
| 0.1 | 1.3 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.1 | 0.4 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.1 | 1.3 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.1 | 0.8 | GO:0031087 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 1.3 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 1.1 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.1 | 0.7 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.1 | 1.0 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.5 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.1 | 1.6 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.2 | GO:1990120 | regulation of nucleobase-containing compound transport(GO:0032239) positive regulation of nucleobase-containing compound transport(GO:0032241) positive regulation of nucleocytoplasmic transport(GO:0046824) regulation of RNA export from nucleus(GO:0046831) positive regulation of RNA export from nucleus(GO:0046833) messenger ribonucleoprotein complex assembly(GO:1990120) |
| 0.1 | 1.3 | GO:0046855 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) inositol phosphate catabolic process(GO:0071545) |
| 0.1 | 1.5 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.1 | 1.3 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.1 | 1.2 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.1 | 0.8 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.1 | 1.3 | GO:0009214 | cyclic nucleotide catabolic process(GO:0009214) |
| 0.0 | 0.5 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.2 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.4 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.3 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.9 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.9 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 0.8 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.7 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.0 | 0.8 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.4 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.0 | 0.1 | GO:0003190 | heart valve formation(GO:0003188) atrioventricular valve formation(GO:0003190) |
| 0.0 | 0.4 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) |
| 0.0 | 1.1 | GO:0034968 | histone lysine methylation(GO:0034968) |
| 0.0 | 0.1 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.0 | 0.5 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.4 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.4 | 1.3 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.4 | 2.9 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.3 | 1.0 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.2 | 1.3 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.1 | 0.8 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 0.6 | GO:0005915 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.1 | 0.9 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 0.7 | GO:0098835 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.1 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 1.0 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 0.5 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 1.5 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 1.2 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.9 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.1 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.1 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.4 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 0.8 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0004113 | 2',3'-cyclic-nucleotide 3'-phosphodiesterase activity(GO:0004113) |
| 0.3 | 0.9 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.3 | 1.3 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.3 | 1.0 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.2 | 0.7 | GO:0061599 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.2 | 0.6 | GO:1990174 | phosphodiesterase decapping endonuclease activity(GO:1990174) |
| 0.2 | 0.6 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.2 | 1.5 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.1 | 0.4 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.1 | 0.6 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.3 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.1 | 1.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 0.9 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 0.4 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 1.5 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 0.3 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.1 | 1.3 | GO:0008252 | nucleotidase activity(GO:0008252) |
| 0.1 | 0.7 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.8 | GO:0019870 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.9 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.4 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.8 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.5 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
| 0.0 | 0.2 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.6 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.5 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.3 | GO:0005223 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.9 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.4 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.2 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.4 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 1.4 | GO:0042054 | histone methyltransferase activity(GO:0042054) |
| 0.0 | 0.6 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.1 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) phosphoprotein binding(GO:0051219) |
| 0.0 | 0.5 | GO:0099604 | ligand-gated calcium channel activity(GO:0099604) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 1.3 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 1.3 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 1.3 | PID IL1 PATHWAY | IL1-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 1.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.3 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.1 | 1.3 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 0.9 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 0.4 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.8 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.9 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 1.0 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.6 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.6 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.9 | REACTOME RECRUITMENT OF MITOTIC CENTROSOME PROTEINS AND COMPLEXES | Genes involved in Recruitment of mitotic centrosome proteins and complexes |
| 0.0 | 1.3 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.0 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 1.3 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |