Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
Results for nkx1.2lb
Z-value: 0.56
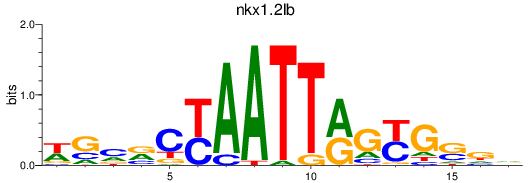
Transcription factors associated with nkx1.2lb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nkx1.2lb
|
ENSDARG00000099427 | NK1 transcription factor related 2-like,b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nkx1.2lb | dr11_v1_chr14_+_15064870_15064870 | -0.80 | 9.1e-03 | Click! |
Activity profile of nkx1.2lb motif
Sorted Z-values of nkx1.2lb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_34002185 | 1.92 |
ENSDART00000046599
|
zar1l
|
zygote arrest 1-like |
| chr8_-_23780334 | 1.24 |
ENSDART00000145179
ENSDART00000145894 |
zgc:195245
|
zgc:195245 |
| chr19_-_18313303 | 1.12 |
ENSDART00000164644
ENSDART00000167480 ENSDART00000163104 |
si:dkey-208k4.2
|
si:dkey-208k4.2 |
| chr2_+_6253246 | 1.10 |
ENSDART00000058256
ENSDART00000076700 |
zp3b
|
zona pellucida glycoprotein 3b |
| chr20_-_14114078 | 1.05 |
ENSDART00000168434
ENSDART00000104032 |
si:ch211-223m11.2
|
si:ch211-223m11.2 |
| chr5_-_3991655 | 1.02 |
ENSDART00000159368
|
myo19
|
myosin XIX |
| chr25_-_32869794 | 0.88 |
ENSDART00000162784
|
tmem266
|
transmembrane protein 266 |
| chr17_-_53329704 | 0.79 |
ENSDART00000193895
|
exd1
|
exonuclease 3'-5' domain containing 1 |
| chr6_+_4255319 | 0.70 |
ENSDART00000170351
|
nbeal1
|
neurobeachin-like 1 |
| chr20_+_14114258 | 0.69 |
ENSDART00000044937
|
kcns3b
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3b |
| chr13_-_6279684 | 0.64 |
ENSDART00000092105
|
agpat5
|
1-acylglycerol-3-phosphate O-acyltransferase 5 (lysophosphatidic acid acyltransferase, epsilon) |
| chr14_-_30945515 | 0.60 |
ENSDART00000161540
|
si:zfos-80g12.1
|
si:zfos-80g12.1 |
| chr25_+_20272145 | 0.59 |
ENSDART00000109605
|
si:dkey-219c3.2
|
si:dkey-219c3.2 |
| chr11_+_31864921 | 0.57 |
ENSDART00000180252
|
diaph3
|
diaphanous-related formin 3 |
| chr7_-_40630698 | 0.52 |
ENSDART00000134547
|
ube3c
|
ubiquitin protein ligase E3C |
| chr1_-_48933 | 0.51 |
ENSDART00000171162
|
ildr1a
|
immunoglobulin-like domain containing receptor 1a |
| chr9_-_50000144 | 0.49 |
ENSDART00000123416
|
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr17_-_15189397 | 0.49 |
ENSDART00000133710
ENSDART00000110507 |
wdhd1
|
WD repeat and HMG-box DNA binding protein 1 |
| chr5_-_67629263 | 0.48 |
ENSDART00000133753
|
zbtb20
|
zinc finger and BTB domain containing 20 |
| chr12_-_4249000 | 0.47 |
ENSDART00000059298
|
zgc:92313
|
zgc:92313 |
| chr21_-_31013817 | 0.47 |
ENSDART00000065504
|
ncbp3
|
nuclear cap binding subunit 3 |
| chr8_-_39822917 | 0.46 |
ENSDART00000067843
|
zgc:162025
|
zgc:162025 |
| chr8_-_20243389 | 0.45 |
ENSDART00000184904
|
acer1
|
alkaline ceramidase 1 |
| chr7_-_59123066 | 0.45 |
ENSDART00000175438
|
dennd4c
|
DENN/MADD domain containing 4C |
| chr13_-_36798204 | 0.45 |
ENSDART00000012357
|
sav1
|
salvador family WW domain containing protein 1 |
| chr13_+_35955562 | 0.42 |
ENSDART00000137377
|
si:ch211-67f13.7
|
si:ch211-67f13.7 |
| chr25_+_36312459 | 0.41 |
ENSDART00000182484
|
CR354435.5
|
|
| chr20_-_37813863 | 0.40 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr23_-_31913069 | 0.40 |
ENSDART00000135526
|
mtfr2
|
mitochondrial fission regulator 2 |
| chr6_-_12275836 | 0.39 |
ENSDART00000189980
|
pkp4
|
plakophilin 4 |
| chr13_-_36663358 | 0.39 |
ENSDART00000085319
|
sos2
|
son of sevenless homolog 2 (Drosophila) |
| chr7_+_20384408 | 0.37 |
ENSDART00000173809
|
si:dkey-33c9.6
|
si:dkey-33c9.6 |
| chr7_-_64971839 | 0.37 |
ENSDART00000164682
|
sinhcafl
|
SIN3-HDAC complex associated factor, like |
| chr2_+_10006839 | 0.37 |
ENSDART00000160304
|
slc35a3b
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3b |
| chr22_+_30089054 | 0.36 |
ENSDART00000185369
|
add3a
|
adducin 3 (gamma) a |
| chr4_+_306036 | 0.34 |
ENSDART00000103659
|
msgn1
|
mesogenin 1 |
| chr11_+_12811906 | 0.34 |
ENSDART00000123445
|
rtel1
|
regulator of telomere elongation helicase 1 |
| chr17_+_21902058 | 0.34 |
ENSDART00000142178
|
plekha1a
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1a |
| chr24_-_29586082 | 0.33 |
ENSDART00000136763
|
vav3a
|
vav 3 guanine nucleotide exchange factor a |
| chr6_+_31368071 | 0.32 |
ENSDART00000038990
|
jak1
|
Janus kinase 1 |
| chr21_-_33995710 | 0.31 |
ENSDART00000100508
ENSDART00000179622 |
ebf1b
|
early B cell factor 1b |
| chr13_-_30142087 | 0.31 |
ENSDART00000110157
|
tysnd1
|
trypsin domain containing 1 |
| chr21_+_4410733 | 0.30 |
ENSDART00000136831
ENSDART00000131612 |
HTRA2 (1 of many)
|
si:dkey-84o3.7 |
| chr24_-_32582378 | 0.29 |
ENSDART00000066590
|
rdh12l
|
retinol dehydrogenase 12, like |
| chr8_+_7756893 | 0.29 |
ENSDART00000191894
|
fgd1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr13_-_9236905 | 0.28 |
ENSDART00000142597
ENSDART00000147176 ENSDART00000181670 |
HTRA2 (1 of many)
si:dkey-112g5.12
|
si:dkey-33c12.12 si:dkey-112g5.12 |
| chr13_+_12761707 | 0.27 |
ENSDART00000015127
|
zgc:100846
|
zgc:100846 |
| chr2_+_10007113 | 0.27 |
ENSDART00000155213
|
slc35a3b
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3b |
| chr5_-_22052852 | 0.27 |
ENSDART00000002938
|
mtmr8
|
myotubularin related protein 8 |
| chr3_-_40836081 | 0.27 |
ENSDART00000143135
|
wipi2
|
WD repeat domain, phosphoinositide interacting 2 |
| chr17_+_16429826 | 0.26 |
ENSDART00000136078
|
efcab11
|
EF-hand calcium binding domain 11 |
| chr8_-_4031121 | 0.26 |
ENSDART00000169474
ENSDART00000163754 |
mtmr3
|
myotubularin related protein 3 |
| chr6_+_23026714 | 0.26 |
ENSDART00000124948
|
srp68
|
signal recognition particle 68 |
| chr2_+_54696042 | 0.26 |
ENSDART00000074270
|
ankrd12
|
ankyrin repeat domain 12 |
| chr5_-_33298352 | 0.26 |
ENSDART00000015076
|
zgc:103692
|
zgc:103692 |
| chr4_+_63790028 | 0.25 |
ENSDART00000160964
|
si:dkey-92j16.2
|
si:dkey-92j16.2 |
| chr13_-_9045879 | 0.25 |
ENSDART00000155463
ENSDART00000140041 ENSDART00000137454 |
si:dkey-112g5.11
|
si:dkey-112g5.11 |
| chr13_+_9468535 | 0.25 |
ENSDART00000135088
ENSDART00000164270 ENSDART00000099619 ENSDART00000164656 |
HTRA2 (1 of many)
|
si:dkey-265c15.6 |
| chr13_-_9257728 | 0.23 |
ENSDART00000145985
ENSDART00000169991 ENSDART00000132764 |
HTRA2 (1 of many)
|
zgc:173425 |
| chr13_-_9111927 | 0.23 |
ENSDART00000133815
ENSDART00000109783 ENSDART00000142540 ENSDART00000192613 |
HTRA2 (1 of many)
si:dkey-112g5.12
|
si:dkey-19p15.4 si:dkey-112g5.12 |
| chr9_+_16241656 | 0.23 |
ENSDART00000154326
|
si:ch211-261p9.4
|
si:ch211-261p9.4 |
| chr13_-_9184132 | 0.22 |
ENSDART00000135289
ENSDART00000136309 ENSDART00000132630 |
HTRA2 (1 of many)
|
si:dkey-33c12.10 |
| chr13_+_9496748 | 0.22 |
ENSDART00000147050
ENSDART00000102120 |
CR848040.4
|
|
| chr21_+_4328348 | 0.22 |
ENSDART00000131658
ENSDART00000143799 |
HTRA2 (1 of many)
|
si:dkey-84o3.7 |
| chr21_-_15200556 | 0.22 |
ENSDART00000141809
|
sfswap
|
splicing factor SWAP |
| chr12_-_19119176 | 0.21 |
ENSDART00000149180
|
aco2
|
aconitase 2, mitochondrial |
| chr13_-_9213207 | 0.20 |
ENSDART00000139861
ENSDART00000140524 |
HTRA2 (1 of many)
|
si:dkey-33c12.11 |
| chr1_-_44812014 | 0.20 |
ENSDART00000139044
|
si:dkey-9i23.8
|
si:dkey-9i23.8 |
| chr13_+_9521629 | 0.19 |
ENSDART00000149870
ENSDART00000137666 |
si:dkey-19p15.4
|
si:dkey-19p15.4 |
| chr3_+_32832538 | 0.19 |
ENSDART00000139410
|
cd2bp2
|
CD2 (cytoplasmic tail) binding protein 2 |
| chr13_-_9159852 | 0.19 |
ENSDART00000170185
ENSDART00000158697 ENSDART00000143393 ENSDART00000164973 ENSDART00000159910 |
HTRA2 (1 of many)
|
si:dkey-112g5.16 |
| chr21_+_4429001 | 0.18 |
ENSDART00000134520
|
HTRA2 (1 of many)
|
si:dkey-84o3.6 |
| chr16_+_32051572 | 0.18 |
ENSDART00000039109
|
leng1
|
leukocyte receptor cluster (LRC) member 1 |
| chr9_+_1313418 | 0.18 |
ENSDART00000191361
|
casp8l2
|
caspase 8, apoptosis-related cysteine peptidase, like 2 |
| chr10_-_105100 | 0.18 |
ENSDART00000145716
|
ttc3
|
tetratricopeptide repeat domain 3 |
| chr2_-_55298075 | 0.17 |
ENSDART00000186404
ENSDART00000149062 |
rab8a
|
RAB8A, member RAS oncogene family |
| chr18_+_13236255 | 0.17 |
ENSDART00000091560
|
tldc1
|
TBC/LysM-associated domain containing 1 |
| chr21_+_4368735 | 0.17 |
ENSDART00000182817
|
CT027677.1
|
|
| chr1_+_55758257 | 0.17 |
ENSDART00000139312
|
tecrb
|
trans-2,3-enoyl-CoA reductase b |
| chr13_-_9070754 | 0.16 |
ENSDART00000143783
ENSDART00000102121 ENSDART00000140820 ENSDART00000184210 |
si:dkey-112g5.12
|
si:dkey-112g5.12 |
| chr21_+_4348600 | 0.16 |
ENSDART00000138463
|
HTRA2 (1 of many)
|
si:dkey-84o3.2 |
| chr7_+_24881680 | 0.16 |
ENSDART00000058843
|
krcp
|
kelch repeat-containing protein |
| chr25_-_7705487 | 0.14 |
ENSDART00000128099
|
prdm11
|
PR domain containing 11 |
| chr23_-_35064785 | 0.14 |
ENSDART00000172240
|
BX294434.1
|
|
| chr16_+_4078240 | 0.14 |
ENSDART00000160890
|
inpp5b
|
inositol polyphosphate-5-phosphatase B |
| chr9_+_35015747 | 0.14 |
ENSDART00000140110
|
gabpa
|
GA binding protein transcription factor, alpha subunit |
| chr7_+_20383841 | 0.13 |
ENSDART00000052906
|
si:dkey-33c9.6
|
si:dkey-33c9.6 |
| chr17_+_24821627 | 0.12 |
ENSDART00000112389
|
wdr43
|
WD repeat domain 43 |
| chr4_-_44611273 | 0.12 |
ENSDART00000156793
|
znf1115
|
zinc finger protein 1115 |
| chr22_-_17458070 | 0.12 |
ENSDART00000139658
|
si:ch211-197g15.10
|
si:ch211-197g15.10 |
| chr9_-_35334642 | 0.12 |
ENSDART00000157195
|
ncam2
|
neural cell adhesion molecule 2 |
| chr20_+_36812368 | 0.08 |
ENSDART00000062931
|
abracl
|
ABRA C-terminal like |
| chr13_+_411607 | 0.07 |
ENSDART00000186483
|
CU570800.3
|
|
| chr2_+_25315591 | 0.06 |
ENSDART00000161386
|
fndc3ba
|
fibronectin type III domain containing 3Ba |
| chr7_+_13418812 | 0.05 |
ENSDART00000191905
ENSDART00000091567 |
dagla
|
diacylglycerol lipase, alpha |
| chr4_-_42039794 | 0.05 |
ENSDART00000110313
|
si:dkeyp-85d8.5
|
si:dkeyp-85d8.5 |
| chr23_-_1348933 | 0.03 |
ENSDART00000168981
|
CABZ01078120.1
|
|
| chr21_-_21570134 | 0.03 |
ENSDART00000174388
|
or133-4
|
odorant receptor, family H, subfamily 133, member 4 |
| chr14_+_918287 | 0.02 |
ENSDART00000167066
|
CU462913.1
|
|
| chr7_+_34453185 | 0.02 |
ENSDART00000173875
ENSDART00000173921 ENSDART00000173995 |
si:cabz01009626.1
|
si:cabz01009626.1 |
| chr2_+_19522082 | 0.02 |
ENSDART00000146098
|
pimr49
|
Pim proto-oncogene, serine/threonine kinase, related 49 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nkx1.2lb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0048340 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) |
| 0.1 | 0.5 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.1 | 1.5 | GO:0035803 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.6 | GO:0071233 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.1 | 0.5 | GO:0097065 | anterior head development(GO:0097065) |
| 0.0 | 0.2 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.0 | 0.6 | GO:0090481 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.8 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.5 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.3 | GO:0036268 | swimming(GO:0036268) |
| 0.0 | 0.3 | GO:0032205 | negative regulation of telomere maintenance(GO:0032205) |
| 0.0 | 0.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:0071869 | response to monoamine(GO:0071867) response to catecholamine(GO:0071869) response to epinephrine(GO:0071871) |
| 0.0 | 0.4 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.3 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 | 0.3 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.4 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.3 | GO:0071594 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.0 | 0.3 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.7 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.0 | GO:0098920 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 0.0 | 0.6 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.6 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 0.5 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:1990923 | PET complex(GO:1990923) |
| 0.1 | 0.6 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.3 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.3 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.6 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.1 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.5 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 0.2 | GO:0005682 | U5 snRNP(GO:0005682) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.1 | 0.6 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 0.3 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 0.5 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.4 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.2 | GO:0003994 | aconitate hydratase activity(GO:0003994) |
| 0.1 | 0.6 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 0.5 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 0.3 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.6 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.0 | 0.5 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.7 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.8 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 0.1 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.0 | 0.3 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.3 | GO:0030165 | PDZ domain binding(GO:0030165) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.4 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |