Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
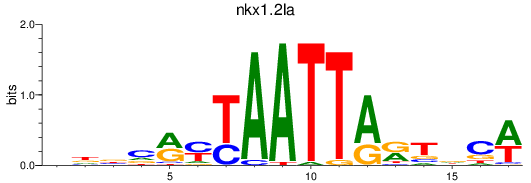
Results for nkx1.2la
Z-value: 1.38
Transcription factors associated with nkx1.2la
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nkx1.2la
|
ENSDARG00000006350 | NK1 transcription factor related 2-like,a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nkx1.2la | dr11_v1_chr13_+_40770628_40770628 | 0.09 | 8.2e-01 | Click! |
Activity profile of nkx1.2la motif
Sorted Z-values of nkx1.2la motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_23984179 | 2.94 |
ENSDART00000175879
|
apoc2
|
apolipoprotein C-II |
| chr4_-_9891874 | 2.94 |
ENSDART00000067193
|
adm2a
|
adrenomedullin 2a |
| chr25_+_29160102 | 2.81 |
ENSDART00000162854
|
pkmb
|
pyruvate kinase M1/2b |
| chr8_+_16025554 | 2.67 |
ENSDART00000110171
|
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr4_+_16885854 | 2.39 |
ENSDART00000017726
|
etnk1
|
ethanolamine kinase 1 |
| chr21_-_43015383 | 2.38 |
ENSDART00000065097
|
dpysl3
|
dihydropyrimidinase-like 3 |
| chr15_+_23799461 | 2.32 |
ENSDART00000154885
|
si:ch211-167j9.4
|
si:ch211-167j9.4 |
| chr16_-_20312146 | 2.12 |
ENSDART00000134980
|
si:dkeyp-86h10.3
|
si:dkeyp-86h10.3 |
| chr23_+_19813677 | 2.11 |
ENSDART00000139192
ENSDART00000142308 |
emd
|
emerin (Emery-Dreifuss muscular dystrophy) |
| chr13_-_9886579 | 2.09 |
ENSDART00000101926
|
si:ch211-117n7.7
|
si:ch211-117n7.7 |
| chr7_+_59020972 | 1.93 |
ENSDART00000157873
|
hacd1
|
3-hydroxyacyl-CoA dehydratase 1 |
| chr4_+_9669717 | 1.85 |
ENSDART00000004604
|
si:dkey-153k10.9
|
si:dkey-153k10.9 |
| chr21_+_6751405 | 1.82 |
ENSDART00000037265
ENSDART00000146371 |
olfm1b
|
olfactomedin 1b |
| chr23_-_15216654 | 1.82 |
ENSDART00000131649
|
sulf2b
|
sulfatase 2b |
| chr21_+_6751760 | 1.81 |
ENSDART00000135914
|
olfm1b
|
olfactomedin 1b |
| chr5_-_51619742 | 1.74 |
ENSDART00000188537
|
otpb
|
orthopedia homeobox b |
| chr13_+_35339182 | 1.68 |
ENSDART00000019323
|
jag1b
|
jagged 1b |
| chr4_+_21129752 | 1.62 |
ENSDART00000169764
|
syt1a
|
synaptotagmin Ia |
| chr20_+_19512727 | 1.61 |
ENSDART00000063696
|
atraid
|
all-trans retinoic acid-induced differentiation factor |
| chr15_-_5815006 | 1.58 |
ENSDART00000102459
|
rbp2a
|
retinol binding protein 2a, cellular |
| chr6_+_57541776 | 1.54 |
ENSDART00000157330
|
necab3
|
N-terminal EF-hand calcium binding protein 3 |
| chr8_-_15129573 | 1.53 |
ENSDART00000142358
|
bcar3
|
BCAR3, NSP family adaptor protein |
| chr25_+_11008419 | 1.50 |
ENSDART00000156589
|
mhc1lia
|
major histocompatibility complex class I LIA |
| chr17_-_49978986 | 1.50 |
ENSDART00000154728
|
col12a1a
|
collagen, type XII, alpha 1a |
| chr8_+_25079470 | 1.49 |
ENSDART00000000744
|
sypl2b
|
synaptophysin-like 2b |
| chr17_-_12385308 | 1.40 |
ENSDART00000080927
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr18_+_21408794 | 1.37 |
ENSDART00000140161
|
necab2
|
N-terminal EF-hand calcium binding protein 2 |
| chr20_+_43925266 | 1.37 |
ENSDART00000037379
|
clic5b
|
chloride intracellular channel 5b |
| chr14_-_413273 | 1.36 |
ENSDART00000163976
ENSDART00000179907 |
FAT4
|
FAT atypical cadherin 4 |
| chr2_-_38000276 | 1.34 |
ENSDART00000034790
|
pcp4l1
|
Purkinje cell protein 4 like 1 |
| chr6_-_7720332 | 1.30 |
ENSDART00000135945
|
rpsa
|
ribosomal protein SA |
| chr7_+_6652967 | 1.28 |
ENSDART00000102681
|
pnp5a
|
purine nucleoside phosphorylase 5a |
| chr24_+_38301080 | 1.26 |
ENSDART00000105672
|
mybpc2b
|
myosin binding protein C, fast type b |
| chr21_+_28478663 | 1.26 |
ENSDART00000077887
ENSDART00000134150 |
slc22a6l
|
solute carrier family 22 (organic anion transporter), member 6, like |
| chr23_+_43255328 | 1.22 |
ENSDART00000102712
|
tgm2a
|
transglutaminase 2, C polypeptide A |
| chr17_+_43032529 | 1.22 |
ENSDART00000055611
ENSDART00000154863 |
isca2
|
iron-sulfur cluster assembly 2 |
| chr10_-_40968095 | 1.20 |
ENSDART00000184104
|
npffr1l1
|
neuropeptide FF receptor 1 like 1 |
| chr8_+_23521974 | 1.19 |
ENSDART00000188130
ENSDART00000129378 |
sema3gb
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Gb |
| chr11_-_30158191 | 1.18 |
ENSDART00000155278
ENSDART00000156121 |
scml2
|
Scm polycomb group protein like 2 |
| chr10_+_37137464 | 1.17 |
ENSDART00000114909
|
cuedc1a
|
CUE domain containing 1a |
| chr5_+_22969651 | 1.16 |
ENSDART00000089992
ENSDART00000145477 |
hmgn7
|
high mobility group nucleosomal binding domain 7 |
| chr23_+_28582865 | 1.15 |
ENSDART00000020296
|
l1cama
|
L1 cell adhesion molecule, paralog a |
| chr20_+_28861435 | 1.13 |
ENSDART00000142678
|
slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter), member 1 |
| chr11_+_30057762 | 1.11 |
ENSDART00000164139
|
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr1_-_50859053 | 1.09 |
ENSDART00000132779
ENSDART00000137648 |
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr17_+_21964472 | 1.04 |
ENSDART00000063704
ENSDART00000188904 |
crip3
|
cysteine-rich protein 3 |
| chr3_-_12930217 | 1.02 |
ENSDART00000166322
|
pdgfab
|
platelet-derived growth factor alpha polypeptide b |
| chr6_+_7421898 | 0.99 |
ENSDART00000043946
|
ccdc65
|
coiled-coil domain containing 65 |
| chr1_+_44127292 | 0.97 |
ENSDART00000160542
|
cabp2a
|
calcium binding protein 2a |
| chr24_-_21689146 | 0.96 |
ENSDART00000105917
|
urad
|
ureidoimidazoline (2-oxo-4-hydroxy-4-carboxy-5-) decarboxylase |
| chr19_+_43780970 | 0.94 |
ENSDART00000063870
|
rpl11
|
ribosomal protein L11 |
| chr15_+_9327252 | 0.94 |
ENSDART00000144381
|
sgcg
|
sarcoglycan, gamma |
| chr14_+_5385855 | 0.92 |
ENSDART00000031508
|
lbx2
|
ladybird homeobox 2 |
| chr20_+_28861629 | 0.91 |
ENSDART00000187274
ENSDART00000047826 |
slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter), member 1 |
| chr4_+_6032640 | 0.89 |
ENSDART00000157487
|
tfec
|
transcription factor EC |
| chr8_+_18588551 | 0.88 |
ENSDART00000177476
ENSDART00000063539 |
prrg1
|
proline rich Gla (G-carboxyglutamic acid) 1 |
| chr24_-_31425799 | 0.87 |
ENSDART00000157998
|
cngb3.1
|
cyclic nucleotide gated channel beta 3, tandem duplicate 1 |
| chr11_-_30634286 | 0.87 |
ENSDART00000191019
|
zgc:153665
|
zgc:153665 |
| chr23_-_20051369 | 0.87 |
ENSDART00000049836
|
bgnb
|
biglycan b |
| chr4_+_9400012 | 0.81 |
ENSDART00000191960
|
tmtc1
|
transmembrane and tetratricopeptide repeat containing 1 |
| chr21_-_19314618 | 0.80 |
ENSDART00000188744
|
gpat3
|
glycerol-3-phosphate acyltransferase 3 |
| chr24_-_36680261 | 0.77 |
ENSDART00000059507
|
ccr10
|
chemokine (C-C motif) receptor 10 |
| chr18_+_24919614 | 0.76 |
ENSDART00000008638
|
rgma
|
repulsive guidance molecule family member a |
| chr15_-_14552101 | 0.75 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr2_+_50608099 | 0.74 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr19_+_12237945 | 0.72 |
ENSDART00000190034
|
grhl2b
|
grainyhead-like transcription factor 2b |
| chr1_+_51721851 | 0.70 |
ENSDART00000040397
|
prdx2
|
peroxiredoxin 2 |
| chr7_+_54642005 | 0.69 |
ENSDART00000171864
|
fgf19
|
fibroblast growth factor 19 |
| chr5_+_1079423 | 0.65 |
ENSDART00000172231
|
si:zfos-128g4.2
|
si:zfos-128g4.2 |
| chr15_+_45640906 | 0.64 |
ENSDART00000149361
ENSDART00000149079 |
sagb
|
S-antigen; retina and pineal gland (arrestin) b |
| chr22_-_26865361 | 0.64 |
ENSDART00000182504
|
hmox2a
|
heme oxygenase 2a |
| chr11_-_21528056 | 0.63 |
ENSDART00000181626
|
srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr16_-_11798994 | 0.63 |
ENSDART00000135408
|
cnfn
|
cornifelin |
| chr25_+_19489370 | 0.62 |
ENSDART00000091132
|
glsl
|
glutaminase like |
| chr8_+_52637507 | 0.61 |
ENSDART00000163830
|
si:dkey-90l8.3
|
si:dkey-90l8.3 |
| chr19_-_5669122 | 0.60 |
ENSDART00000112211
|
si:ch211-264f5.2
|
si:ch211-264f5.2 |
| chr12_+_695619 | 0.60 |
ENSDART00000161691
|
abcc3
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
| chr22_+_9939901 | 0.58 |
ENSDART00000169777
ENSDART00000081420 |
zgc:171686
|
zgc:171686 |
| chr10_-_5581487 | 0.56 |
ENSDART00000141943
|
syk
|
spleen tyrosine kinase |
| chr11_+_16153207 | 0.55 |
ENSDART00000192356
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr7_+_34620418 | 0.55 |
ENSDART00000081338
|
slc9a5
|
solute carrier family 9, subfamily A (NHE5, cation proton antiporter 5), member 5 |
| chr4_+_5255041 | 0.53 |
ENSDART00000137966
|
ccdc167
|
coiled-coil domain containing 167 |
| chr18_+_49969568 | 0.52 |
ENSDART00000126916
|
mob2b
|
MOB kinase activator 2b |
| chr1_-_19502322 | 0.50 |
ENSDART00000181888
ENSDART00000044030 |
kitb
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog b |
| chr20_-_13774826 | 0.50 |
ENSDART00000063831
|
opn8c
|
opsin 8, group member c |
| chr6_+_28051978 | 0.49 |
ENSDART00000143218
|
si:ch73-194h10.2
|
si:ch73-194h10.2 |
| chr16_+_14033121 | 0.48 |
ENSDART00000135844
|
rusc1
|
RUN and SH3 domain containing 1 |
| chr11_-_29737088 | 0.46 |
ENSDART00000159828
|
si:ch211-218g23.6
|
si:ch211-218g23.6 |
| chr20_-_42102416 | 0.46 |
ENSDART00000186378
ENSDART00000188253 ENSDART00000186458 |
slc35f1
|
solute carrier family 35, member F1 |
| chr4_-_45115963 | 0.46 |
ENSDART00000137248
|
si:dkey-51d8.6
|
si:dkey-51d8.6 |
| chr22_+_7738966 | 0.45 |
ENSDART00000147073
|
si:ch73-44m9.5
|
si:ch73-44m9.5 |
| chr22_-_8725768 | 0.45 |
ENSDART00000189873
ENSDART00000181819 |
si:ch73-27e22.1
si:ch73-27e22.8
|
si:ch73-27e22.1 si:ch73-27e22.8 |
| chr21_+_16145980 | 0.44 |
ENSDART00000135119
|
qrfpr4
|
pyroglutamylated RFamide peptide receptor 4 |
| chr4_+_57583139 | 0.41 |
ENSDART00000158802
|
il17ra1b
|
interleukin 17 receptor A1b |
| chr14_+_30795559 | 0.41 |
ENSDART00000006132
|
cfl1
|
cofilin 1 |
| chr10_-_5847655 | 0.41 |
ENSDART00000192773
|
ankrd55
|
ankyrin repeat domain 55 |
| chr8_+_18011522 | 0.40 |
ENSDART00000136756
|
ssbp3b
|
single stranded DNA binding protein 3b |
| chr3_-_39180048 | 0.39 |
ENSDART00000049720
|
cdk21
|
cyclin-dependent kinase 21 |
| chr5_-_25723079 | 0.39 |
ENSDART00000014013
|
gda
|
guanine deaminase |
| chr1_-_19215336 | 0.37 |
ENSDART00000162949
ENSDART00000170680 |
ptprdb
|
protein tyrosine phosphatase, receptor type, D, b |
| chr10_+_35358675 | 0.37 |
ENSDART00000193263
|
si:dkey-259j3.5
|
si:dkey-259j3.5 |
| chr23_-_1660708 | 0.37 |
ENSDART00000175138
|
CU693481.1
|
|
| chr5_-_68074592 | 0.35 |
ENSDART00000165052
ENSDART00000018792 |
spag7
|
sperm associated antigen 7 |
| chr1_+_33668236 | 0.34 |
ENSDART00000122316
ENSDART00000102184 |
arl13b
|
ADP-ribosylation factor-like 13b |
| chr2_+_53204750 | 0.34 |
ENSDART00000163644
|
zgc:165603
|
zgc:165603 |
| chr8_-_53044089 | 0.33 |
ENSDART00000135982
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr6_+_52877147 | 0.33 |
ENSDART00000174370
|
or137-8
|
odorant receptor, family H, subfamily 137, member 8 |
| chr22_-_35330532 | 0.33 |
ENSDART00000172654
|
FO818743.1
|
|
| chr14_+_36521005 | 0.31 |
ENSDART00000192286
|
TENM3
|
si:dkey-237h12.3 |
| chr9_+_43799829 | 0.29 |
ENSDART00000186240
|
ube2e3
|
ubiquitin-conjugating enzyme E2E 3 (UBC4/5 homolog, yeast) |
| chr11_-_16152400 | 0.29 |
ENSDART00000123665
|
arpc4l
|
actin related protein 2/3 complex, subunit 4, like |
| chr6_-_40842768 | 0.29 |
ENSDART00000076160
|
mustn1a
|
musculoskeletal, embryonic nuclear protein 1a |
| chr8_+_28695914 | 0.28 |
ENSDART00000033386
|
ocstamp
|
osteoclast stimulatory transmembrane protein |
| chr11_-_2838699 | 0.27 |
ENSDART00000066189
|
lhfpl5a
|
LHFPL tetraspan subfamily member 5a |
| chr25_+_20715950 | 0.27 |
ENSDART00000180223
|
ergic2
|
ERGIC and golgi 2 |
| chr9_-_41062412 | 0.26 |
ENSDART00000193879
|
ankar
|
ankyrin and armadillo repeat containing |
| chr23_+_16815353 | 0.25 |
ENSDART00000142789
ENSDART00000143424 |
zgc:100832
|
zgc:100832 |
| chr15_-_35134918 | 0.24 |
ENSDART00000157196
|
si:ch211-272b8.7
|
si:ch211-272b8.7 |
| chr22_-_20011476 | 0.23 |
ENSDART00000093312
ENSDART00000093310 |
celf5a
|
cugbp, Elav-like family member 5a |
| chr18_-_16924221 | 0.22 |
ENSDART00000122102
|
wee1
|
WEE1 G2 checkpoint kinase |
| chr12_-_41684729 | 0.22 |
ENSDART00000184461
|
jakmip3
|
Janus kinase and microtubule interacting protein 3 |
| chr8_-_13184989 | 0.22 |
ENSDART00000135738
|
zgc:194990
|
zgc:194990 |
| chr23_-_7674902 | 0.22 |
ENSDART00000185612
ENSDART00000180524 |
plagl2
|
pleiomorphic adenoma gene-like 2 |
| chr20_-_9095105 | 0.21 |
ENSDART00000140792
|
oma1
|
OMA1 zinc metallopeptidase |
| chr1_-_31171242 | 0.21 |
ENSDART00000190294
|
kcnq5b
|
potassium voltage-gated channel, KQT-like subfamily, member 5b |
| chr16_-_27677930 | 0.20 |
ENSDART00000145991
|
tbrg4
|
transforming growth factor beta regulator 4 |
| chr18_+_17827149 | 0.20 |
ENSDART00000190237
ENSDART00000189345 |
ZNF423
|
si:ch211-216l23.1 |
| chr11_+_2506516 | 0.20 |
ENSDART00000130886
ENSDART00000189767 |
NABP2
|
si:ch73-190f16.2 |
| chr20_-_47188966 | 0.19 |
ENSDART00000152965
|
si:dkeyp-104f11.9
|
si:dkeyp-104f11.9 |
| chr17_-_10122204 | 0.18 |
ENSDART00000160751
|
BX088587.1
|
|
| chr1_-_33351780 | 0.17 |
ENSDART00000178996
|
gyg2
|
glycogenin 2 |
| chr22_-_26865181 | 0.17 |
ENSDART00000138311
|
hmox2a
|
heme oxygenase 2a |
| chr14_-_4556896 | 0.16 |
ENSDART00000044678
ENSDART00000192863 |
GABRA2
|
gamma-aminobutyric acid type A receptor alpha2 subunit |
| chr4_+_20566371 | 0.16 |
ENSDART00000127576
|
BX248410.1
|
|
| chr1_-_17715493 | 0.16 |
ENSDART00000133027
|
si:dkey-256e7.8
|
si:dkey-256e7.8 |
| chr8_-_50888806 | 0.16 |
ENSDART00000053750
|
acsl2
|
acyl-CoA synthetase long chain family member 2 |
| chr18_+_7073130 | 0.16 |
ENSDART00000101216
ENSDART00000148947 |
si:dkey-88e18.2
|
si:dkey-88e18.2 |
| chr7_-_50410524 | 0.15 |
ENSDART00000083346
|
hypk
|
huntingtin interacting protein K |
| chr15_+_15856178 | 0.15 |
ENSDART00000080338
|
dusp14
|
dual specificity phosphatase 14 |
| chr8_-_25034411 | 0.14 |
ENSDART00000135973
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr4_+_11458078 | 0.13 |
ENSDART00000037600
|
ankrd16
|
ankyrin repeat domain 16 |
| chr11_+_16152316 | 0.12 |
ENSDART00000081054
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr12_+_30706158 | 0.10 |
ENSDART00000133869
|
kcnk1a
|
potassium channel, subfamily K, member 1a |
| chr23_-_37575030 | 0.10 |
ENSDART00000031875
|
tor1l3
|
torsin family 1 like 3 |
| chr3_+_54744069 | 0.10 |
ENSDART00000134958
ENSDART00000114443 |
si:ch211-74m13.3
|
si:ch211-74m13.3 |
| chr11_+_705727 | 0.09 |
ENSDART00000165366
|
timp4.2
|
TIMP metallopeptidase inhibitor 4, tandem duplicate 2 |
| chr13_-_37229245 | 0.09 |
ENSDART00000140923
|
si:dkeyp-77c8.5
|
si:dkeyp-77c8.5 |
| chr8_+_25034544 | 0.06 |
ENSDART00000123300
|
ngrn
|
neugrin, neurite outgrowth associated |
| chr16_+_35916371 | 0.06 |
ENSDART00000167208
|
sh3d21
|
SH3 domain containing 21 |
| chr4_-_61691066 | 0.06 |
ENSDART00000159114
|
znf1129
|
zinc finger protein 1129 |
| chr7_-_66868543 | 0.05 |
ENSDART00000149680
|
ampd3a
|
adenosine monophosphate deaminase 3a |
| chr19_+_46158078 | 0.05 |
ENSDART00000183933
ENSDART00000164055 |
cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr3_-_36701870 | 0.04 |
ENSDART00000183457
|
myh11b
|
myosin, heavy chain 11b, smooth muscle |
| chr15_-_47115787 | 0.03 |
ENSDART00000192601
|
CABZ01079081.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of nkx1.2la
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.1 | GO:0006660 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.4 | 2.9 | GO:0042983 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.4 | 1.6 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.3 | 1.0 | GO:0019628 | urate catabolic process(GO:0019628) urate metabolic process(GO:0046415) |
| 0.3 | 1.7 | GO:0021767 | mammillary body development(GO:0021767) |
| 0.2 | 1.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.2 | 2.4 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.2 | 0.6 | GO:0045579 | positive regulation of B cell differentiation(GO:0045579) |
| 0.2 | 0.9 | GO:1904105 | positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.2 | 2.9 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.2 | 0.8 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 2.0 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 0.4 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.1 | 0.8 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.1 | 0.4 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 0.1 | 0.6 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.1 | 0.5 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.1 | 1.0 | GO:0003352 | regulation of cilium movement(GO:0003352) |
| 0.1 | 0.3 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to ciliary membrane(GO:1903441) |
| 0.1 | 1.0 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.1 | 1.7 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 1.5 | GO:0035860 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.1 | 1.4 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.8 | GO:0046341 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 1.5 | GO:0048679 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.1 | 0.9 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.1 | 1.9 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.1 | 0.6 | GO:0006543 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.1 | 1.6 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 0.7 | GO:0060561 | apoptotic process involved in morphogenesis(GO:0060561) |
| 0.1 | 0.7 | GO:1990748 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.1 | 1.5 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.1 | 0.3 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.1 | 1.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 1.2 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 0.6 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.0 | 1.1 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.5 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.9 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 2.8 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 0.3 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.2 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.0 | 0.2 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.0 | 2.1 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.7 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.7 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 0.1 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 1.2 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.7 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.2 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.4 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.3 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 1.2 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.6 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.3 | 2.9 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.2 | 1.4 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.1 | 0.9 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 6.3 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 1.0 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.0 | 0.7 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.8 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.6 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.6 | GO:0044327 | dendritic spine head(GO:0044327) phagocytic vesicle(GO:0045335) |
| 0.0 | 1.3 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.4 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 2.0 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 1.5 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 1.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.3 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.3 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.9 | GO:0018812 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.5 | 2.8 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.3 | 2.0 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.2 | 1.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.2 | 2.4 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.2 | 2.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 1.3 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 0.8 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 1.0 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 0.8 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 0.7 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 1.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 2.9 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.6 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 0.9 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.1 | 1.7 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 1.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.8 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 1.3 | GO:0043236 | laminin binding(GO:0043236) |
| 0.1 | 1.2 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.5 | GO:0015386 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.8 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 1.6 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.4 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.4 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 1.6 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.6 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.7 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 1.2 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 1.0 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.8 | GO:0019957 | chemokine binding(GO:0019956) C-C chemokine binding(GO:0019957) |
| 0.0 | 1.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 2.9 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.0 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.2 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 1.4 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.7 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.5 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.8 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 1.3 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.5 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.6 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.8 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.7 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.5 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.9 | PID CMYB PATHWAY | C-MYB transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.6 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.3 | 2.9 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 2.4 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.2 | 2.4 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.2 | 0.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 0.7 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 0.4 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.1 | 0.4 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.6 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 1.3 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.8 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.7 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.9 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |