Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
Results for nfkb1_rela
Z-value: 1.10
Transcription factors associated with nfkb1_rela
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
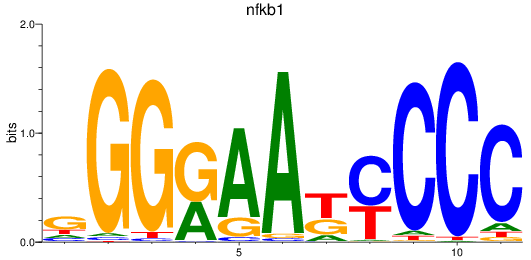
nfkb1
|
ENSDARG00000105261 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
|
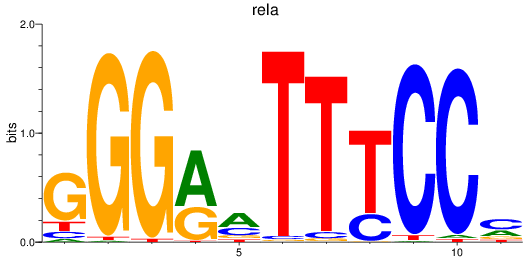
rela
|
ENSDARG00000098696 | v-rel avian reticuloendotheliosis viral oncogene homolog A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rela | dr11_v1_chr7_-_804515_804515 | -0.90 | 1.0e-03 | Click! |
| nfkb1 | dr11_v1_chr14_-_49859747_49859823 | -0.75 | 1.9e-02 | Click! |
Activity profile of nfkb1_rela motif
Sorted Z-values of nfkb1_rela motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_55131289 | 3.42 |
ENSDART00000111585
|
AL935210.1
|
|
| chr3_+_55114097 | 2.70 |
ENSDART00000121686
|
hbbe1.1
|
hemoglobin beta embryonic-1.1 |
| chr2_+_42191592 | 2.50 |
ENSDART00000144716
|
cavin4a
|
caveolae associated protein 4a |
| chr10_-_39011514 | 1.80 |
ENSDART00000075123
|
pcp4a
|
Purkinje cell protein 4a |
| chr21_+_28958471 | 1.72 |
ENSDART00000144331
ENSDART00000005929 |
ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr16_-_50175069 | 1.66 |
ENSDART00000192979
|
lim2.5
|
lens intrinsic membrane protein 2.5 |
| chr15_+_32727848 | 1.58 |
ENSDART00000161361
|
postnb
|
periostin, osteoblast specific factor b |
| chr24_-_33703504 | 1.55 |
ENSDART00000079292
|
cavin4b
|
caveolae associated protein 4b |
| chr11_-_5865744 | 1.36 |
ENSDART00000104360
|
gamt
|
guanidinoacetate N-methyltransferase |
| chr20_+_28434196 | 1.35 |
ENSDART00000034245
|
dpf3
|
D4, zinc and double PHD fingers, family 3 |
| chr19_+_19767567 | 1.35 |
ENSDART00000169074
|
hoxa3a
|
homeobox A3a |
| chr3_+_55122662 | 1.35 |
ENSDART00000128380
|
hbbe1.2
|
hemoglobin beta embryonic-1.2 |
| chr22_-_651719 | 1.34 |
ENSDART00000148692
|
apobec2a
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2a |
| chr23_+_31815423 | 1.25 |
ENSDART00000075730
ENSDART00000075726 |
myb
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr25_+_37397031 | 1.24 |
ENSDART00000193643
ENSDART00000169132 |
slc1a2b
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2b |
| chr23_+_36115541 | 1.22 |
ENSDART00000130090
|
hoxc6a
|
homeobox C6a |
| chr2_+_2737422 | 1.20 |
ENSDART00000032459
|
aqp1a.1
|
aquaporin 1a (Colton blood group), tandem duplicate 1 |
| chr15_-_24869826 | 1.15 |
ENSDART00000127047
|
tusc5a
|
tumor suppressor candidate 5a |
| chr20_-_20533865 | 1.13 |
ENSDART00000125039
|
six6b
|
SIX homeobox 6b |
| chr7_-_28696556 | 1.12 |
ENSDART00000148822
|
adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr3_-_28120092 | 1.04 |
ENSDART00000151143
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr12_+_7445595 | 1.03 |
ENSDART00000103536
ENSDART00000152524 |
phyhiplb
|
phytanoyl-CoA 2-hydroxylase interacting protein-like b |
| chr2_+_16780643 | 1.02 |
ENSDART00000125647
ENSDART00000108611 ENSDART00000181245 ENSDART00000163194 |
tfa
|
transferrin-a |
| chr25_-_27843066 | 1.01 |
ENSDART00000179684
ENSDART00000186000 ENSDART00000190065 |
asb15a
|
ankyrin repeat and SOCS box containing 15a |
| chr1_+_18716 | 1.00 |
ENSDART00000172454
ENSDART00000161190 |
nfkbiz
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
| chr6_-_58757131 | 1.00 |
ENSDART00000083582
|
soat2
|
sterol O-acyltransferase 2 |
| chr3_+_46459540 | 0.99 |
ENSDART00000188150
|
si:ch211-66e2.5
|
si:ch211-66e2.5 |
| chr15_+_27364394 | 0.96 |
ENSDART00000122101
|
tbx2b
|
T-box 2b |
| chr6_-_10305918 | 0.95 |
ENSDART00000090994
|
scn1lab
|
sodium channel, voltage-gated, type I like, alpha b |
| chr6_-_54815886 | 0.94 |
ENSDART00000180793
ENSDART00000007498 |
tnni1b
|
troponin I type 1b (skeletal, slow) |
| chr22_-_16180467 | 0.93 |
ENSDART00000171331
ENSDART00000185607 |
vcam1b
|
vascular cell adhesion molecule 1b |
| chr13_-_2010191 | 0.91 |
ENSDART00000161021
ENSDART00000124134 |
gfral
|
GDNF family receptor alpha like |
| chr13_+_51579851 | 0.91 |
ENSDART00000163847
|
nkx6.2
|
NK6 homeobox 2 |
| chr6_-_607063 | 0.90 |
ENSDART00000189900
|
lgals2b
|
lectin, galactoside-binding, soluble, 2b |
| chr7_-_34262080 | 0.89 |
ENSDART00000183246
|
si:ch211-98n17.5
|
si:ch211-98n17.5 |
| chr9_-_98982 | 0.86 |
ENSDART00000147882
|
lims2
|
LIM and senescent cell antigen-like domains 2 |
| chr13_+_15004398 | 0.86 |
ENSDART00000057810
|
emx1
|
empty spiracles homeobox 1 |
| chr4_-_25485404 | 0.86 |
ENSDART00000041402
|
pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr9_+_993477 | 0.86 |
ENSDART00000182045
|
CABZ01066921.1
|
|
| chr12_+_26471712 | 0.83 |
ENSDART00000162115
|
ndel1a
|
nudE neurodevelopment protein 1-like 1a |
| chr21_+_30937690 | 0.83 |
ENSDART00000022562
|
rhogb
|
ras homolog family member Gb |
| chr16_+_23403602 | 0.82 |
ENSDART00000159848
|
s100w
|
S100 calcium binding protein W |
| chr1_+_16397063 | 0.80 |
ENSDART00000159794
|
micu3a
|
mitochondrial calcium uptake family, member 3a |
| chr18_+_27738349 | 0.80 |
ENSDART00000187816
|
tspan18b
|
tetraspanin 18b |
| chr20_+_16881883 | 0.80 |
ENSDART00000130107
|
nfkbiaa
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha a |
| chr8_+_25302172 | 0.78 |
ENSDART00000046182
ENSDART00000145316 |
gstm.3
|
glutathione S-transferase mu tandem duplicate 3 |
| chr1_+_16396870 | 0.76 |
ENSDART00000189245
ENSDART00000113266 |
micu3a
|
mitochondrial calcium uptake family, member 3a |
| chr4_-_22311610 | 0.76 |
ENSDART00000137814
|
hcls1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr14_+_36885524 | 0.73 |
ENSDART00000032547
|
lect2l
|
leukocyte cell-derived chemotaxin 2 like |
| chr16_-_16590780 | 0.73 |
ENSDART00000059841
|
si:ch211-257p13.3
|
si:ch211-257p13.3 |
| chr16_+_32995882 | 0.72 |
ENSDART00000170157
|
prss35
|
protease, serine, 35 |
| chr11_-_35763323 | 0.72 |
ENSDART00000026017
|
bhlhe40
|
basic helix-loop-helix family, member e40 |
| chr4_-_149334 | 0.72 |
ENSDART00000163280
|
tbk1
|
TANK-binding kinase 1 |
| chr17_+_8175998 | 0.71 |
ENSDART00000131200
|
myct1b
|
myc target 1b |
| chr1_+_33328857 | 0.71 |
ENSDART00000137151
|
mxra5a
|
matrix-remodelling associated 5a |
| chr5_-_66702479 | 0.71 |
ENSDART00000129197
|
mn1b
|
meningioma 1b |
| chr2_-_42234484 | 0.71 |
ENSDART00000132617
ENSDART00000136690 ENSDART00000141358 |
apom
|
apolipoprotein M |
| chr14_-_4121052 | 0.70 |
ENSDART00000167074
|
irf2
|
interferon regulatory factor 2 |
| chr3_-_21242460 | 0.69 |
ENSDART00000007293
|
tcap
|
titin-cap (telethonin) |
| chr8_-_38477817 | 0.69 |
ENSDART00000075989
|
inpp5l
|
inositol polyphosphate-5-phosphatase L |
| chr10_-_27196093 | 0.69 |
ENSDART00000185282
|
auts2a
|
autism susceptibility candidate 2a |
| chr11_+_8129536 | 0.67 |
ENSDART00000158112
ENSDART00000011183 |
prkacba
|
protein kinase, cAMP-dependent, catalytic, beta a |
| chr25_-_35960229 | 0.67 |
ENSDART00000073434
|
snx20
|
sorting nexin 20 |
| chr12_+_34732262 | 0.67 |
ENSDART00000169634
|
si:dkey-21c1.8
|
si:dkey-21c1.8 |
| chr4_-_25485214 | 0.67 |
ENSDART00000159941
|
pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr1_+_33322555 | 0.67 |
ENSDART00000113486
|
mxra5a
|
matrix-remodelling associated 5a |
| chr12_+_22576404 | 0.65 |
ENSDART00000172053
|
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr8_+_42998944 | 0.65 |
ENSDART00000048819
|
rassf2a
|
Ras association (RalGDS/AF-6) domain family member 2a |
| chr12_-_30760971 | 0.65 |
ENSDART00000066257
|
entpd1
|
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr16_+_13855039 | 0.64 |
ENSDART00000113764
ENSDART00000143983 |
zgc:174888
|
zgc:174888 |
| chr25_+_5288665 | 0.64 |
ENSDART00000169540
|
CABZ01039861.1
|
|
| chr2_-_30200206 | 0.64 |
ENSDART00000130142
|
ube2w
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr19_+_19412692 | 0.62 |
ENSDART00000113580
|
wu:fc38h03
|
wu:fc38h03 |
| chr6_-_436658 | 0.62 |
ENSDART00000191515
|
grap2b
|
GRB2-related adaptor protein 2b |
| chr9_-_18716 | 0.59 |
ENSDART00000164763
|
CABZ01078737.1
|
|
| chr5_+_37032038 | 0.58 |
ENSDART00000045036
|
nfkbib
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr17_-_51195651 | 0.57 |
ENSDART00000191205
ENSDART00000088185 |
paplna
|
papilin a, proteoglycan-like sulfated glycoprotein |
| chr6_+_2030703 | 0.57 |
ENSDART00000109679
ENSDART00000187502 ENSDART00000191165 ENSDART00000187544 |
quo
|
quattro |
| chr23_-_12423778 | 0.57 |
ENSDART00000124091
|
wfdc2
|
WAP four-disulfide core domain 2 |
| chr24_-_4973765 | 0.56 |
ENSDART00000127597
|
zic1
|
zic family member 1 (odd-paired homolog, Drosophila) |
| chr23_-_43609595 | 0.56 |
ENSDART00000172222
|
CABZ01117603.1
|
|
| chr8_-_13029297 | 0.56 |
ENSDART00000144305
|
dennd2da
|
DENN/MADD domain containing 2Da |
| chr11_-_396724 | 0.55 |
ENSDART00000184375
|
CR855375.3
|
|
| chr11_+_2156430 | 0.55 |
ENSDART00000182896
|
hoxc13b
|
homeobox C13b |
| chr25_+_10458990 | 0.54 |
ENSDART00000130354
ENSDART00000044738 |
ric8a
|
RIC8 guanine nucleotide exchange factor A |
| chr8_+_24281512 | 0.53 |
ENSDART00000062845
|
mmp9
|
matrix metallopeptidase 9 |
| chr11_-_32723851 | 0.53 |
ENSDART00000155592
|
pcdh17
|
protocadherin 17 |
| chr23_-_37835794 | 0.53 |
ENSDART00000137358
|
cd40
|
CD40 molecule, TNF receptor superfamily member 5 |
| chr14_+_33722950 | 0.52 |
ENSDART00000075312
|
apln
|
apelin |
| chr18_-_18543358 | 0.52 |
ENSDART00000126460
|
il34
|
interleukin 34 |
| chr5_+_21100356 | 0.51 |
ENSDART00000051586
|
hs3st1l2
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 1-like 2 |
| chr3_-_58733718 | 0.51 |
ENSDART00000154603
|
si:ch73-281f12.4
|
si:ch73-281f12.4 |
| chr2_+_6963296 | 0.50 |
ENSDART00000147146
|
ddr2b
|
discoidin domain receptor tyrosine kinase 2b |
| chr13_-_31452516 | 0.50 |
ENSDART00000193268
|
rtn1a
|
reticulon 1a |
| chr10_+_7029664 | 0.50 |
ENSDART00000166206
|
psd3l
|
pleckstrin and Sec7 domain containing 3, like |
| chr14_-_7888748 | 0.49 |
ENSDART00000166293
|
ppp3cb
|
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr16_-_35975254 | 0.49 |
ENSDART00000167537
|
eva1ba
|
eva-1 homolog Ba (C. elegans) |
| chr14_-_30967284 | 0.48 |
ENSDART00000149435
|
il2rgb
|
interleukin 2 receptor, gamma b |
| chr12_+_5081759 | 0.48 |
ENSDART00000164178
|
prrt2
|
proline-rich transmembrane protein 2 |
| chr9_+_26103814 | 0.48 |
ENSDART00000026011
|
efnb2a
|
ephrin-B2a |
| chr13_-_45795722 | 0.48 |
ENSDART00000141779
|
fndc5a
|
fibronectin type III domain containing 5a |
| chr9_-_6380653 | 0.47 |
ENSDART00000078523
|
ecrg4a
|
esophageal cancer related gene 4a |
| chr14_-_31080183 | 0.47 |
ENSDART00000173282
|
mbnl3
|
muscleblind-like splicing regulator 3 |
| chr13_+_12739283 | 0.47 |
ENSDART00000102279
|
lingo2b
|
leucine rich repeat and Ig domain containing 2b |
| chr25_-_6557854 | 0.46 |
ENSDART00000181740
|
cspg4
|
chondroitin sulfate proteoglycan 4 |
| chr13_-_22843562 | 0.46 |
ENSDART00000142738
|
pbld
|
phenazine biosynthesis like protein domain containing |
| chr12_+_27462225 | 0.46 |
ENSDART00000105661
|
meox1
|
mesenchyme homeobox 1 |
| chr7_+_39634873 | 0.46 |
ENSDART00000114774
|
ptpn5
|
protein tyrosine phosphatase, non-receptor type 5 |
| chr7_+_57677120 | 0.46 |
ENSDART00000110623
|
arsj
|
arylsulfatase family, member J |
| chr5_-_64168415 | 0.44 |
ENSDART00000048395
|
cmlc1
|
cardiac myosin light chain-1 |
| chr21_+_5862476 | 0.44 |
ENSDART00000065848
|
wdr38
|
WD repeat domain 38 |
| chr13_-_39947335 | 0.44 |
ENSDART00000056996
|
sfrp5
|
secreted frizzled-related protein 5 |
| chr20_+_218886 | 0.43 |
ENSDART00000002661
|
lama4
|
laminin, alpha 4 |
| chr7_-_20338048 | 0.43 |
ENSDART00000125594
|
zgc:194312
|
zgc:194312 |
| chr23_+_42819221 | 0.43 |
ENSDART00000180495
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr4_+_3389866 | 0.42 |
ENSDART00000153834
|
sypl1
|
synaptophysin-like 1 |
| chr24_+_22731228 | 0.42 |
ENSDART00000146733
|
si:dkey-225k4.1
|
si:dkey-225k4.1 |
| chr13_+_32148338 | 0.42 |
ENSDART00000188591
|
osr1
|
odd-skipped related transciption factor 1 |
| chr20_+_22045089 | 0.42 |
ENSDART00000063564
ENSDART00000187013 ENSDART00000161552 ENSDART00000174478 ENSDART00000063568 ENSDART00000152247 |
nmu
|
neuromedin U |
| chr17_+_20205185 | 0.42 |
ENSDART00000167007
|
gnrh3
|
gonadotropin-releasing hormone 3 |
| chr15_-_33640890 | 0.41 |
ENSDART00000167705
|
stard13b
|
StAR-related lipid transfer (START) domain containing 13b |
| chr12_+_2381213 | 0.41 |
ENSDART00000188007
|
LO018238.1
|
|
| chr14_-_4120636 | 0.41 |
ENSDART00000059230
|
irf2
|
interferon regulatory factor 2 |
| chr7_+_39758714 | 0.41 |
ENSDART00000111278
|
sorcs2
|
sortilin-related VPS10 domain containing receptor 2 |
| chr25_+_19489370 | 0.41 |
ENSDART00000091132
|
glsl
|
glutaminase like |
| chr13_+_27316934 | 0.41 |
ENSDART00000164533
|
eef1a1a
|
eukaryotic translation elongation factor 1 alpha 1a |
| chr8_+_22355909 | 0.41 |
ENSDART00000146457
ENSDART00000142883 |
zgc:153631
|
zgc:153631 |
| chr2_-_9857932 | 0.41 |
ENSDART00000154490
|
si:ch211-252f13.5
|
si:ch211-252f13.5 |
| chr13_-_33822550 | 0.41 |
ENSDART00000143703
|
flrt3
|
fibronectin leucine rich transmembrane 3 |
| chr9_+_31752628 | 0.40 |
ENSDART00000060054
|
itgbl1
|
integrin, beta-like 1 |
| chr4_-_12914163 | 0.40 |
ENSDART00000140002
ENSDART00000145917 ENSDART00000141355 ENSDART00000067135 |
msrb3
|
methionine sulfoxide reductase B3 |
| chr14_+_36889893 | 0.40 |
ENSDART00000124159
|
si:ch211-132p1.3
|
si:ch211-132p1.3 |
| chr16_+_24681177 | 0.40 |
ENSDART00000058956
ENSDART00000189335 |
ywhabl
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide like |
| chr7_+_22585447 | 0.40 |
ENSDART00000149144
|
chrnb1l
|
cholinergic receptor, nicotinic, beta 1 (muscle) like |
| chr13_-_24311628 | 0.40 |
ENSDART00000004420
|
rab4a
|
RAB4a, member RAS oncogene family |
| chr14_+_36246726 | 0.39 |
ENSDART00000105602
|
elovl6
|
ELOVL fatty acid elongase 6 |
| chr3_-_58650057 | 0.39 |
ENSDART00000057640
|
dhrs7ca
|
dehydrogenase/reductase (SDR family) member 7Ca |
| chr11_-_236984 | 0.38 |
ENSDART00000170778
|
dusp7
|
dual specificity phosphatase 7 |
| chr10_+_31244619 | 0.38 |
ENSDART00000145562
ENSDART00000184412 |
robo4
|
roundabout, axon guidance receptor, homolog 4 (Drosophila) |
| chr19_-_43659779 | 0.38 |
ENSDART00000144058
|
si:ch211-193k19.2
|
si:ch211-193k19.2 |
| chr11_-_42106310 | 0.37 |
ENSDART00000164635
|
si:ch211-193l2.10
|
si:ch211-193l2.10 |
| chr9_-_12624622 | 0.37 |
ENSDART00000146535
|
tspearb
|
thrombospondin-type laminin G domain and EAR repeats b |
| chr24_-_12689571 | 0.37 |
ENSDART00000015517
|
pdcd6
|
programmed cell death 6 |
| chr6_-_18992896 | 0.37 |
ENSDART00000170228
|
sept9b
|
septin 9b |
| chr7_-_46781614 | 0.36 |
ENSDART00000173891
|
tshz3b
|
teashirt zinc finger homeobox 3b |
| chr23_+_3591690 | 0.36 |
ENSDART00000180822
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr5_-_41841675 | 0.36 |
ENSDART00000141683
|
si:dkey-65b12.6
|
si:dkey-65b12.6 |
| chr7_-_15413019 | 0.36 |
ENSDART00000172147
|
si:ch211-106j24.1
|
si:ch211-106j24.1 |
| chr24_+_80653 | 0.35 |
ENSDART00000158473
ENSDART00000129135 |
reck
|
reversion-inducing-cysteine-rich protein with kazal motifs |
| chr25_+_3347461 | 0.35 |
ENSDART00000104888
|
slc35b4
|
solute carrier family 35, member B4 |
| chr7_-_35083585 | 0.35 |
ENSDART00000192732
|
agrp
|
agouti related neuropeptide |
| chr1_+_44919221 | 0.35 |
ENSDART00000140402
|
wu:fc21g02
|
wu:fc21g02 |
| chr6_+_48664275 | 0.35 |
ENSDART00000161184
|
FAM19A3
|
si:dkey-238f9.1 |
| chr3_-_39695856 | 0.35 |
ENSDART00000148247
|
b9d1
|
B9 protein domain 1 |
| chr3_-_49815223 | 0.34 |
ENSDART00000181208
|
gcgra
|
glucagon receptor a |
| chr14_+_49382180 | 0.33 |
ENSDART00000158329
|
tnip1
|
TNFAIP3 interacting protein 1 |
| chr9_+_56103173 | 0.33 |
ENSDART00000172063
|
edar
|
ectodysplasin A receptor |
| chr11_+_8565828 | 0.33 |
ENSDART00000126523
|
tbl1xr1a
|
transducin (beta)-like 1 X-linked receptor 1a |
| chr21_-_44009169 | 0.33 |
ENSDART00000028960
|
ndufa2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2 |
| chr24_-_38816725 | 0.32 |
ENSDART00000063231
|
nog2
|
noggin 2 |
| chr4_+_13810811 | 0.32 |
ENSDART00000067168
|
pdzrn4
|
PDZ domain containing ring finger 4 |
| chr25_+_8955530 | 0.32 |
ENSDART00000156444
|
si:ch211-256a21.4
|
si:ch211-256a21.4 |
| chr1_-_30039331 | 0.32 |
ENSDART00000086935
ENSDART00000143800 |
MARCH4 (1 of many)
|
zgc:153256 |
| chr1_+_32528097 | 0.32 |
ENSDART00000128317
|
nlgn4a
|
neuroligin 4a |
| chr21_+_22845317 | 0.31 |
ENSDART00000065555
|
birc2
|
baculoviral IAP repeat containing 2 |
| chr22_+_110158 | 0.31 |
ENSDART00000143698
|
prkar2ab
|
protein kinase, cAMP-dependent, regulatory, type II, alpha, B |
| chr6_+_58915889 | 0.31 |
ENSDART00000083628
|
ddit3
|
DNA-damage-inducible transcript 3 |
| chr12_+_48634927 | 0.30 |
ENSDART00000168441
|
zgc:165653
|
zgc:165653 |
| chr3_-_52644737 | 0.30 |
ENSDART00000126180
|
si:dkey-210j14.3
|
si:dkey-210j14.3 |
| chr10_-_14536399 | 0.30 |
ENSDART00000186501
|
hdhd2
|
haloacid dehalogenase-like hydrolase domain containing 2 |
| chr7_-_32833153 | 0.30 |
ENSDART00000099871
ENSDART00000099872 |
slc17a6b
|
solute carrier family 17 (vesicular glutamate transporter), member 6b |
| chr15_-_1962326 | 0.30 |
ENSDART00000179669
|
dock10
|
dedicator of cytokinesis 10 |
| chr23_+_45579497 | 0.29 |
ENSDART00000110381
|
egr4
|
early growth response 4 |
| chr13_-_9318891 | 0.29 |
ENSDART00000137364
|
si:dkey-33c12.3
|
si:dkey-33c12.3 |
| chr13_+_35339182 | 0.29 |
ENSDART00000019323
|
jag1b
|
jagged 1b |
| chr5_+_9246018 | 0.29 |
ENSDART00000081769
ENSDART00000183189 |
susd1
|
sushi domain containing 1 |
| chr17_+_38262408 | 0.29 |
ENSDART00000017493
|
nkx2.1
|
NK2 homeobox 1 |
| chr23_+_3616224 | 0.29 |
ENSDART00000190917
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr4_+_73672430 | 0.29 |
ENSDART00000174310
ENSDART00000150505 |
si:dkey-262g12.7
|
si:dkey-262g12.7 |
| chr13_-_27916439 | 0.28 |
ENSDART00000139081
ENSDART00000087097 |
ogfrl1
|
opioid growth factor receptor-like 1 |
| chr18_+_2563756 | 0.28 |
ENSDART00000164147
|
p2ry2.1
|
purinergic receptor P2Y2, tandem duplicate 1 |
| chr17_+_1063988 | 0.28 |
ENSDART00000160400
|
gchfr
|
GTP cyclohydrolase I feedback regulator |
| chr1_+_12135129 | 0.28 |
ENSDART00000126020
|
spink4
|
serine peptidase inhibitor, Kazal type 4 |
| chr16_+_50006872 | 0.28 |
ENSDART00000157100
|
ube2e2
|
ubiquitin-conjugating enzyme E2E 2 |
| chr1_-_37377509 | 0.28 |
ENSDART00000113542
|
tnip2
|
TNFAIP3 interacting protein 2 |
| chr15_+_32821392 | 0.28 |
ENSDART00000158272
|
dclk1b
|
doublecortin-like kinase 1b |
| chr23_+_10347851 | 0.27 |
ENSDART00000127667
|
krt18
|
keratin 18 |
| chr23_-_437467 | 0.27 |
ENSDART00000192106
|
tspan2b
|
tetraspanin 2b |
| chr1_-_36152131 | 0.27 |
ENSDART00000182113
ENSDART00000182904 |
znf827
|
zinc finger protein 827 |
| chr5_+_30608057 | 0.27 |
ENSDART00000133583
|
si:ch211-117m20.4
|
si:ch211-117m20.4 |
| chr9_-_32604414 | 0.27 |
ENSDART00000088876
ENSDART00000166502 |
satb2
|
SATB homeobox 2 |
| chr22_+_661711 | 0.26 |
ENSDART00000113795
|
elf3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr20_-_8321280 | 0.26 |
ENSDART00000083906
ENSDART00000186477 |
dab1a
|
Dab, reelin signal transducer, homolog 1a (Drosophila) |
| chr9_+_41156818 | 0.25 |
ENSDART00000105764
ENSDART00000147052 |
stat4
|
signal transducer and activator of transcription 4 |
| chr17_+_42027969 | 0.25 |
ENSDART00000147563
|
kiz
|
kizuna centrosomal protein |
| chr7_+_15871408 | 0.25 |
ENSDART00000014572
|
pax6b
|
paired box 6b |
| chr19_+_22216778 | 0.25 |
ENSDART00000052521
|
nfatc1
|
nuclear factor of activated T cells 1 |
| chr4_+_20263097 | 0.25 |
ENSDART00000138820
|
lrtm2a
|
leucine-rich repeats and transmembrane domains 2a |
| chr4_+_7888047 | 0.25 |
ENSDART00000104676
|
camk1da
|
calcium/calmodulin-dependent protein kinase 1Da |
| chr16_+_26766423 | 0.25 |
ENSDART00000048036
|
gem
|
GTP binding protein overexpressed in skeletal muscle |
Network of associatons between targets according to the STRING database.
First level regulatory network of nfkb1_rela
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0006600 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 0.4 | 1.3 | GO:0043388 | positive regulation of DNA binding(GO:0043388) |
| 0.4 | 1.2 | GO:0003091 | renal water homeostasis(GO:0003091) renal water transport(GO:0003097) |
| 0.3 | 1.0 | GO:0072025 | distal convoluted tubule development(GO:0072025) late distal convoluted tubule development(GO:0072068) |
| 0.3 | 1.0 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.3 | 2.2 | GO:0033292 | T-tubule organization(GO:0033292) |
| 0.3 | 0.8 | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation(GO:1901533) |
| 0.3 | 0.5 | GO:0045649 | regulation of macrophage differentiation(GO:0045649) |
| 0.2 | 1.1 | GO:0070445 | regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.2 | 1.2 | GO:0030852 | regulation of granulocyte differentiation(GO:0030852) regulation of neutrophil differentiation(GO:0045658) |
| 0.2 | 3.1 | GO:0048016 | inositol phosphate-mediated signaling(GO:0048016) |
| 0.2 | 1.3 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.2 | 1.3 | GO:0035777 | pronephric distal tubule development(GO:0035777) |
| 0.2 | 0.7 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.2 | 0.5 | GO:0002631 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 0.2 | 0.5 | GO:0045191 | response to protozoan(GO:0001562) TRIF-dependent toll-like receptor signaling pathway(GO:0035666) defense response to protozoan(GO:0042832) regulation of isotype switching(GO:0045191) positive regulation of isotype switching(GO:0045830) |
| 0.2 | 0.5 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.1 | 0.4 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.1 | 1.6 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.1 | 0.8 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 2.7 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 0.8 | GO:0090133 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.1 | 2.0 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.1 | 0.4 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.1 | 0.4 | GO:1905048 | regulation of metallopeptidase activity(GO:1905048) |
| 0.1 | 1.0 | GO:0034433 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.1 | 0.9 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 0.6 | GO:0033986 | response to methanol(GO:0033986) cellular response to methanol(GO:0071405) |
| 0.1 | 0.4 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 0.5 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.1 | 1.5 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 1.6 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 0.3 | GO:0034134 | toll-like receptor 2 signaling pathway(GO:0034134) |
| 0.1 | 1.4 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.1 | 0.2 | GO:0035971 | peptidyl-histidine dephosphorylation(GO:0035971) |
| 0.1 | 0.6 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 0.1 | 0.3 | GO:0060546 | negative regulation of necroptotic process(GO:0060546) negative regulation of necrotic cell death(GO:0060547) |
| 0.1 | 0.3 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.1 | 0.2 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) semicircular canal fusion(GO:0060879) |
| 0.1 | 0.4 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.1 | 0.3 | GO:0070255 | regulation of mucus secretion(GO:0070255) positive regulation of mucus secretion(GO:0070257) |
| 0.1 | 0.4 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 0.2 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.1 | 0.4 | GO:0006543 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.0 | 0.2 | GO:1901842 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 1.2 | GO:0008345 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.0 | 0.5 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.0 | 1.0 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.0 | 0.4 | GO:2000253 | adult feeding behavior(GO:0008343) positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.1 | GO:0072679 | thymocyte migration(GO:0072679) |
| 0.0 | 0.2 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.2 | GO:0048939 | anterior lateral line nerve glial cell differentiation(GO:0048913) myelination of anterior lateral line nerve axons(GO:0048914) anterior lateral line nerve glial cell development(GO:0048939) anterior lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048940) |
| 0.0 | 0.7 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.3 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.3 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.0 | 1.1 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.0 | 0.2 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) |
| 0.0 | 0.1 | GO:0006867 | asparagine transport(GO:0006867) |
| 0.0 | 0.4 | GO:2000406 | positive regulation of lymphocyte migration(GO:2000403) positive regulation of T cell migration(GO:2000406) |
| 0.0 | 0.5 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.4 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.0 | 0.8 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 1.3 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.0 | 2.7 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.1 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.0 | 0.4 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.0 | 0.2 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.0 | 0.7 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.4 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.8 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.3 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.4 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.2 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.2 | GO:0071678 | olfactory bulb axon guidance(GO:0071678) |
| 0.0 | 0.2 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.6 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.9 | GO:0060048 | cardiac muscle contraction(GO:0060048) |
| 0.0 | 0.1 | GO:0090467 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.3 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 0.2 | GO:0042119 | granulocyte activation(GO:0036230) neutrophil activation(GO:0042119) |
| 0.0 | 0.5 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 0.1 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.0 | 0.2 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.4 | GO:0001649 | osteoblast differentiation(GO:0001649) |
| 0.0 | 0.2 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.0 | 0.7 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.2 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 0.0 | 0.5 | GO:0007259 | JAK-STAT cascade(GO:0007259) STAT cascade(GO:0097696) |
| 0.0 | 0.1 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.2 | 2.7 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 1.6 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 4.1 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 0.4 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 1.3 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 1.0 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.4 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.0 | 0.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.2 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 0.1 | GO:0070743 | interleukin-12 complex(GO:0043514) interleukin-23 complex(GO:0070743) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 1.0 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.3 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 1.0 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.7 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.9 | GO:0001726 | ruffle(GO:0001726) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.2 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.3 | 0.8 | GO:0031704 | apelin receptor binding(GO:0031704) |
| 0.2 | 1.0 | GO:0004772 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.2 | 2.7 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.2 | 1.3 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.2 | 0.7 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.2 | 1.2 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.1 | 0.4 | GO:0031530 | gonadotropin hormone-releasing hormone activity(GO:0005183) gonadotropin-releasing hormone receptor binding(GO:0031530) |
| 0.1 | 0.7 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 1.5 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 0.9 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.4 | GO:0031782 | type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.1 | 0.7 | GO:0052659 | inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.1 | 0.4 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.1 | 0.3 | GO:0016519 | gastric inhibitory peptide receptor activity(GO:0016519) |
| 0.1 | 0.4 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.1 | 0.4 | GO:0004984 | olfactory receptor activity(GO:0004984) odorant binding(GO:0005549) |
| 0.1 | 0.2 | GO:0101006 | protein histidine phosphatase activity(GO:0101006) |
| 0.1 | 0.3 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.1 | 0.5 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 0.5 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.0 | 0.2 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.4 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.3 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 1.0 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.5 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.2 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.0 | 0.6 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.3 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.2 | GO:0038132 | neuregulin receptor activity(GO:0038131) neuregulin binding(GO:0038132) |
| 0.0 | 0.5 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.8 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.9 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.7 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.3 | GO:0043028 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 0.2 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.1 | GO:0042164 | interleukin-12 binding(GO:0019972) interleukin-12 alpha subunit binding(GO:0042164) |
| 0.0 | 0.3 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 0.1 | GO:0015182 | L-histidine transmembrane transporter activity(GO:0005290) L-asparagine transmembrane transporter activity(GO:0015182) |
| 0.0 | 0.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.2 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 1.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.3 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.4 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.4 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.2 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.5 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.0 | 0.5 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.5 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.2 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.2 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.4 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 1.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.4 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.2 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.2 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 0.5 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.2 | 2.5 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 2.2 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.5 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 1.2 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.4 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.9 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.3 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.4 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.6 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.3 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.3 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 1.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 0.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 1.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.4 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.5 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.5 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.4 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.7 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.4 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.1 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |