Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
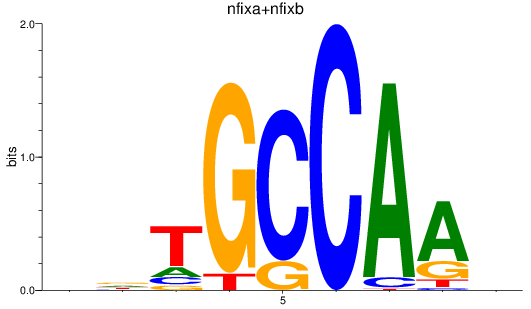
Results for nfixa+nfixb
Z-value: 0.92
Transcription factors associated with nfixa+nfixb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nfixa
|
ENSDARG00000043226 | nuclear factor I/Xa |
|
nfixb
|
ENSDARG00000061836 | nuclear factor I/Xb |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nfixa | dr11_v1_chr1_+_51903413_51903413 | -0.87 | 2.3e-03 | Click! |
| nfixb | dr11_v1_chr3_+_14768364_14768364 | -0.19 | 6.2e-01 | Click! |
Activity profile of nfixa+nfixb motif
Sorted Z-values of nfixa+nfixb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_55139127 | 2.46 |
ENSDART00000115324
|
hbae1.3
|
hemoglobin, alpha embryonic 1.3 |
| chr12_-_16595177 | 1.66 |
ENSDART00000133962
|
si:dkey-239j18.2
|
si:dkey-239j18.2 |
| chr13_+_17672527 | 1.53 |
ENSDART00000148269
ENSDART00000137776 |
comtd1
|
catechol-O-methyltransferase domain containing 1 |
| chr2_+_38161318 | 1.52 |
ENSDART00000044264
|
mmp14b
|
matrix metallopeptidase 14b (membrane-inserted) |
| chr9_-_33107237 | 1.52 |
ENSDART00000013918
|
casq2
|
calsequestrin 2 |
| chr19_-_7225060 | 1.49 |
ENSDART00000133179
ENSDART00000151138 ENSDART00000104799 ENSDART00000128331 |
col11a2
|
collagen, type XI, alpha 2 |
| chr15_-_29388012 | 1.47 |
ENSDART00000115032
|
serpinh1b
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1b |
| chr8_-_52715911 | 1.45 |
ENSDART00000168241
|
tubb2b
|
tubulin, beta 2b |
| chr13_+_30804367 | 1.44 |
ENSDART00000053946
|
cxcl12a
|
chemokine (C-X-C motif) ligand 12a (stromal cell-derived factor 1) |
| chr12_-_17712393 | 1.43 |
ENSDART00000143534
ENSDART00000010144 |
pvalb2
|
parvalbumin 2 |
| chr22_-_15602593 | 1.37 |
ENSDART00000036075
|
tpm4a
|
tropomyosin 4a |
| chr16_-_55028740 | 1.33 |
ENSDART00000156368
ENSDART00000161704 |
zgc:114181
|
zgc:114181 |
| chr5_+_37087583 | 1.21 |
ENSDART00000049900
|
tagln2
|
transgelin 2 |
| chr14_+_18785727 | 1.21 |
ENSDART00000184452
|
si:ch211-111e20.1
|
si:ch211-111e20.1 |
| chr21_+_26390549 | 1.20 |
ENSDART00000185643
|
tmsb
|
thymosin, beta |
| chr11_-_11575070 | 1.18 |
ENSDART00000142208
|
zgc:110712
|
zgc:110712 |
| chr16_+_23961276 | 1.18 |
ENSDART00000192754
|
apoeb
|
apolipoprotein Eb |
| chr17_+_7595356 | 1.17 |
ENSDART00000130625
|
si:dkeyp-110a12.4
|
si:dkeyp-110a12.4 |
| chr22_-_15602760 | 1.17 |
ENSDART00000009054
|
tpm4a
|
tropomyosin 4a |
| chr15_+_36966369 | 1.16 |
ENSDART00000163622
|
kirrel3l
|
kirre like nephrin family adhesion molecule 3, like |
| chr4_+_25680480 | 1.14 |
ENSDART00000100737
|
acot17
|
acyl-CoA thioesterase 17 |
| chr5_-_32309129 | 1.13 |
ENSDART00000123003
|
myhz1.1
|
myosin, heavy polypeptide 1.1, skeletal muscle |
| chr14_+_11457500 | 1.11 |
ENSDART00000169202
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr16_-_13595027 | 1.11 |
ENSDART00000060004
|
ntd5
|
ntl-dependent gene 5 |
| chr11_-_21030070 | 1.10 |
ENSDART00000186322
|
fmoda
|
fibromodulin a |
| chr7_-_8416750 | 1.06 |
ENSDART00000181857
|
jac1
|
jacalin 1 |
| chr14_+_11458044 | 1.05 |
ENSDART00000186425
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr5_+_53482597 | 1.03 |
ENSDART00000180333
|
BX323994.1
|
|
| chr15_-_29387446 | 1.02 |
ENSDART00000145976
ENSDART00000035096 |
serpinh1b
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1b |
| chr16_+_23960933 | 1.01 |
ENSDART00000146077
|
apoeb
|
apolipoprotein Eb |
| chr14_+_25465346 | 1.01 |
ENSDART00000173436
|
si:dkey-280e21.3
|
si:dkey-280e21.3 |
| chr23_+_44741500 | 1.00 |
ENSDART00000166421
|
atp1b2a
|
ATPase Na+/K+ transporting subunit beta 2a |
| chr5_+_20693724 | 0.97 |
ENSDART00000141368
|
si:ch211-240b21.2
|
si:ch211-240b21.2 |
| chr14_-_21064199 | 0.96 |
ENSDART00000172099
|
si:dkey-74k8.3
|
si:dkey-74k8.3 |
| chr22_+_5478353 | 0.95 |
ENSDART00000160596
|
tppp
|
tubulin polymerization promoting protein |
| chr6_-_24301324 | 0.93 |
ENSDART00000171401
|
BX927081.1
|
|
| chr20_+_26880668 | 0.92 |
ENSDART00000077769
|
serpinb1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
| chr11_+_44579865 | 0.91 |
ENSDART00000173425
|
nid1b
|
nidogen 1b |
| chr1_+_1789357 | 0.90 |
ENSDART00000006449
|
atp1a1a.2
|
ATPase Na+/K+ transporting subunit alpha 1a, tandem duplicate 2 |
| chr17_+_33719415 | 0.87 |
ENSDART00000132294
|
fut8a
|
fucosyltransferase 8a (alpha (1,6) fucosyltransferase) |
| chr24_+_2519761 | 0.85 |
ENSDART00000106619
|
nrn1a
|
neuritin 1a |
| chr13_-_9895564 | 0.84 |
ENSDART00000169831
ENSDART00000142629 |
si:ch211-117n7.6
|
si:ch211-117n7.6 |
| chr16_-_22713152 | 0.83 |
ENSDART00000140953
ENSDART00000143836 |
si:ch211-105c13.3
|
si:ch211-105c13.3 |
| chr3_-_24980067 | 0.82 |
ENSDART00000048871
|
desi1a
|
desumoylating isopeptidase 1a |
| chr22_+_38767780 | 0.80 |
ENSDART00000149499
|
alpi.1
|
alkaline phosphatase, intestinal, tandem duplicate 1 |
| chr21_-_4032650 | 0.80 |
ENSDART00000151648
|
ntng2b
|
netrin g2b |
| chr22_+_5176693 | 0.77 |
ENSDART00000160927
|
cers1
|
ceramide synthase 1 |
| chr16_-_45209684 | 0.77 |
ENSDART00000184595
|
fxyd1
|
FXYD domain containing ion transport regulator 1 (phospholemman) |
| chr15_-_18361475 | 0.76 |
ENSDART00000155866
|
zbtb16b
|
zinc finger and BTB domain containing 16b |
| chr14_+_6954579 | 0.75 |
ENSDART00000060998
|
nme5
|
NME/NM23 family member 5 |
| chr5_-_29570141 | 0.74 |
ENSDART00000043259
|
entpd2a.2
|
ectonucleoside triphosphate diphosphohydrolase 2a, tandem duplicate 2 |
| chr18_+_3037998 | 0.72 |
ENSDART00000185844
ENSDART00000162657 |
rps3
|
ribosomal protein S3 |
| chr22_+_38229321 | 0.72 |
ENSDART00000132670
ENSDART00000104504 |
si:ch211-284e20.8
|
si:ch211-284e20.8 |
| chr19_-_42556086 | 0.72 |
ENSDART00000051731
|
si:dkey-267n13.1
|
si:dkey-267n13.1 |
| chr7_+_26132665 | 0.71 |
ENSDART00000129834
|
nat16
|
N-acetyltransferase 16 |
| chr3_-_32603191 | 0.71 |
ENSDART00000150997
|
si:ch73-248e21.7
|
si:ch73-248e21.7 |
| chr22_+_5176255 | 0.70 |
ENSDART00000092647
|
cers1
|
ceramide synthase 1 |
| chr10_+_39263827 | 0.70 |
ENSDART00000172509
|
foxred1
|
FAD-dependent oxidoreductase domain containing 1 |
| chr18_+_44649804 | 0.69 |
ENSDART00000059063
|
ehd2b
|
EH-domain containing 2b |
| chr20_+_539852 | 0.69 |
ENSDART00000185994
|
dse
|
dermatan sulfate epimerase |
| chr22_+_28446365 | 0.68 |
ENSDART00000189359
|
abi3bpb
|
ABI family, member 3 (NESH) binding protein b |
| chr22_-_11729350 | 0.67 |
ENSDART00000105813
|
krt222
|
keratin 222 |
| chr7_+_59649399 | 0.67 |
ENSDART00000123520
ENSDART00000040771 |
rpl34
|
ribosomal protein L34 |
| chr2_+_31382590 | 0.66 |
ENSDART00000086863
|
colec12
|
collectin sub-family member 12 |
| chr18_-_48983690 | 0.66 |
ENSDART00000182359
|
FO681288.3
|
|
| chr4_+_17279966 | 0.64 |
ENSDART00000067005
ENSDART00000137487 |
bcat1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr3_+_26044969 | 0.64 |
ENSDART00000133523
|
hmgxb4a
|
HMG box domain containing 4a |
| chr19_+_30387999 | 0.63 |
ENSDART00000145396
|
tspan13b
|
tetraspanin 13b |
| chr9_-_6927587 | 0.63 |
ENSDART00000059092
|
tmem182a
|
transmembrane protein 182a |
| chr20_+_40237441 | 0.62 |
ENSDART00000168928
|
si:ch211-199i15.5
|
si:ch211-199i15.5 |
| chr13_-_33683889 | 0.61 |
ENSDART00000136820
ENSDART00000065435 |
cst3
|
cystatin C (amyloid angiopathy and cerebral hemorrhage) |
| chr1_+_36651059 | 0.61 |
ENSDART00000187475
|
ednraa
|
endothelin receptor type Aa |
| chr12_+_26471712 | 0.60 |
ENSDART00000162115
|
ndel1a
|
nudE neurodevelopment protein 1-like 1a |
| chr22_+_11857356 | 0.59 |
ENSDART00000179540
|
mras
|
muscle RAS oncogene homolog |
| chr3_-_58644920 | 0.59 |
ENSDART00000155953
|
dhrs7ca
|
dehydrogenase/reductase (SDR family) member 7Ca |
| chr10_-_17988779 | 0.59 |
ENSDART00000132206
ENSDART00000144841 |
si:dkey-242g16.2
|
si:dkey-242g16.2 |
| chr14_-_7128980 | 0.59 |
ENSDART00000171311
|
si:ch73-43g23.1
|
si:ch73-43g23.1 |
| chr22_-_23253252 | 0.57 |
ENSDART00000175556
|
lhx9
|
LIM homeobox 9 |
| chr13_+_18545819 | 0.57 |
ENSDART00000101859
ENSDART00000110197 ENSDART00000136064 |
zgc:154058
|
zgc:154058 |
| chr8_+_20880848 | 0.56 |
ENSDART00000134488
ENSDART00000138605 ENSDART00000192234 |
si:ch73-196i15.3
|
si:ch73-196i15.3 |
| chr18_-_40481028 | 0.55 |
ENSDART00000134177
|
zgc:101040
|
zgc:101040 |
| chr8_-_22965214 | 0.55 |
ENSDART00000148178
|
emilin3a
|
elastin microfibril interfacer 3a |
| chr6_-_55958705 | 0.55 |
ENSDART00000155963
|
eya2
|
EYA transcriptional coactivator and phosphatase 2 |
| chr24_+_26432541 | 0.54 |
ENSDART00000105784
ENSDART00000122554 |
cldn11b
|
claudin 11b |
| chr24_-_2917540 | 0.54 |
ENSDART00000164776
|
fam69c
|
family with sequence similarity 69, member C |
| chr19_-_24443867 | 0.54 |
ENSDART00000163763
ENSDART00000043133 |
thbs3b
|
thrombospondin 3b |
| chr5_-_26181863 | 0.53 |
ENSDART00000098500
|
ccdc125
|
coiled-coil domain containing 125 |
| chr7_-_32833153 | 0.53 |
ENSDART00000099871
ENSDART00000099872 |
slc17a6b
|
solute carrier family 17 (vesicular glutamate transporter), member 6b |
| chr20_+_32497260 | 0.53 |
ENSDART00000145175
ENSDART00000132921 |
si:ch73-257c13.2
|
si:ch73-257c13.2 |
| chr6_-_21873266 | 0.52 |
ENSDART00000151658
ENSDART00000151152 ENSDART00000151179 |
si:dkey-19e4.5
|
si:dkey-19e4.5 |
| chr5_-_10239079 | 0.51 |
ENSDART00000132739
|
si:ch73-42k18.1
|
si:ch73-42k18.1 |
| chr5_-_68916455 | 0.51 |
ENSDART00000171465
|
ank1a
|
ankyrin 1, erythrocytic a |
| chr5_-_51830997 | 0.50 |
ENSDART00000163616
|
homer1b
|
homer scaffolding protein 1b |
| chr7_-_30174882 | 0.50 |
ENSDART00000110409
|
frmd5
|
FERM domain containing 5 |
| chr23_+_13721826 | 0.47 |
ENSDART00000142494
|
zbtb46
|
zinc finger and BTB domain containing 46 |
| chr7_-_43857938 | 0.47 |
ENSDART00000183602
|
cdh11
|
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chr3_-_22228602 | 0.46 |
ENSDART00000017750
|
myl4
|
myosin, light chain 4, alkali; atrial, embryonic |
| chr18_+_26750516 | 0.46 |
ENSDART00000110843
|
alpk3a
|
alpha-kinase 3a |
| chr4_+_37406676 | 0.45 |
ENSDART00000130981
|
si:ch73-134f24.1
|
si:ch73-134f24.1 |
| chr11_+_23704410 | 0.44 |
ENSDART00000112655
|
nfasca
|
neurofascin homolog (chicken) a |
| chr11_-_3865472 | 0.44 |
ENSDART00000161426
|
gata2a
|
GATA binding protein 2a |
| chr14_-_9277152 | 0.44 |
ENSDART00000189048
|
asb12b
|
ankyrin repeat and SOCS box-containing 12b |
| chr9_+_22080122 | 0.42 |
ENSDART00000065956
ENSDART00000136014 |
crygm2e
|
crystallin, gamma M2e |
| chr11_-_3629201 | 0.41 |
ENSDART00000136577
ENSDART00000132121 |
itih3a
|
inter-alpha-trypsin inhibitor heavy chain 3a |
| chr2_-_42375275 | 0.41 |
ENSDART00000026339
|
gtpbp4
|
GTP binding protein 4 |
| chr4_-_35900331 | 0.41 |
ENSDART00000164413
|
zgc:174653
|
zgc:174653 |
| chr1_+_19933065 | 0.41 |
ENSDART00000161573
|
apbb2b
|
amyloid beta (A4) precursor protein-binding, family B, member 2b |
| chr2_-_58193381 | 0.40 |
ENSDART00000159040
|
pnp5b
|
purine nucleoside phosphorylase 5b |
| chr19_-_47455944 | 0.39 |
ENSDART00000190005
|
tfap2e
|
transcription factor AP-2 epsilon |
| chr1_-_9988840 | 0.39 |
ENSDART00000144565
|
si:dkeyp-75b4.9
|
si:dkeyp-75b4.9 |
| chr11_-_22372072 | 0.38 |
ENSDART00000065996
|
tmem183a
|
transmembrane protein 183A |
| chr16_-_28876479 | 0.38 |
ENSDART00000047862
|
si:dkey-239n17.4
|
si:dkey-239n17.4 |
| chr19_+_10831362 | 0.38 |
ENSDART00000053325
|
tomm40l
|
translocase of outer mitochondrial membrane 40 homolog, like |
| chr7_+_35238234 | 0.38 |
ENSDART00000178732
|
tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr5_-_57641257 | 0.38 |
ENSDART00000149282
|
hspb2
|
heat shock protein, alpha-crystallin-related, b2 |
| chr15_+_41836104 | 0.37 |
ENSDART00000137434
|
pxylp1
|
2-phosphoxylose phosphatase 1 |
| chr3_-_32541033 | 0.37 |
ENSDART00000151476
ENSDART00000055324 |
rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr20_+_38525567 | 0.37 |
ENSDART00000147787
|
znf512
|
zinc finger protein 512 |
| chr12_-_37941733 | 0.37 |
ENSDART00000130167
|
CABZ01046949.1
|
|
| chr3_-_5664123 | 0.35 |
ENSDART00000145866
|
si:ch211-106h11.1
|
si:ch211-106h11.1 |
| chr2_-_24488652 | 0.34 |
ENSDART00000052067
|
insl3
|
insulin-like 3 (Leydig cell) |
| chr23_-_3674443 | 0.34 |
ENSDART00000134830
ENSDART00000057422 |
pacsin1a
|
protein kinase C and casein kinase substrate in neurons 1a |
| chr8_+_28469054 | 0.34 |
ENSDART00000062716
|
slc52a3
|
solute carrier family 52 (riboflavin transporter), member 3 |
| chr6_+_32497493 | 0.33 |
ENSDART00000184819
|
kank4
|
KN motif and ankyrin repeat domains 4 |
| chr2_-_14798295 | 0.33 |
ENSDART00000143430
ENSDART00000145869 |
si:ch73-366i20.1
|
si:ch73-366i20.1 |
| chr5_+_53009083 | 0.32 |
ENSDART00000157836
|
hint2
|
histidine triad nucleotide binding protein 2 |
| chr19_-_29832876 | 0.32 |
ENSDART00000005119
|
eif3i
|
eukaryotic translation initiation factor 3, subunit I |
| chr6_+_40661703 | 0.32 |
ENSDART00000142492
|
eno1b
|
enolase 1b, (alpha) |
| chr16_+_38394371 | 0.31 |
ENSDART00000137954
|
cd83
|
CD83 molecule |
| chr17_+_14784275 | 0.31 |
ENSDART00000005355
|
rtraf
|
RNA transcription, translation and transport factor |
| chr4_+_70556298 | 0.31 |
ENSDART00000170985
|
si:dkey-11d20.1
|
si:dkey-11d20.1 |
| chr20_-_15132151 | 0.31 |
ENSDART00000063884
|
si:dkey-239i20.4
|
si:dkey-239i20.4 |
| chr5_+_26121393 | 0.31 |
ENSDART00000002221
|
bco2l
|
beta-carotene 15, 15-dioxygenase 2, like |
| chr7_-_25049377 | 0.30 |
ENSDART00000077191
|
arl2
|
ADP-ribosylation factor-like 2 |
| chr10_-_26738209 | 0.30 |
ENSDART00000188590
|
fgf13b
|
fibroblast growth factor 13b |
| chr19_+_2275019 | 0.30 |
ENSDART00000136138
|
itgb8
|
integrin, beta 8 |
| chr25_-_16581233 | 0.30 |
ENSDART00000155312
|
smc1b
|
structural maintenance of chromosomes 1B |
| chr7_+_57108823 | 0.29 |
ENSDART00000184943
ENSDART00000055956 |
enosf1
|
enolase superfamily member 1 |
| chr1_-_22851481 | 0.29 |
ENSDART00000054386
|
qdprb1
|
quinoid dihydropteridine reductase b1 |
| chr23_+_21414005 | 0.29 |
ENSDART00000144686
|
iffo2a
|
intermediate filament family orphan 2a |
| chr8_+_30671060 | 0.29 |
ENSDART00000193749
|
adora2aa
|
adenosine A2a receptor a |
| chr1_-_9644630 | 0.29 |
ENSDART00000123725
ENSDART00000161164 |
ugt5b3
|
UDP glucuronosyltransferase 5 family, polypeptide B3 |
| chr2_-_51757328 | 0.29 |
ENSDART00000189286
|
si:ch211-9d9.1
|
si:ch211-9d9.1 |
| chr23_+_5524247 | 0.28 |
ENSDART00000189679
ENSDART00000083622 |
tead3a
|
TEA domain family member 3 a |
| chr24_+_14527935 | 0.28 |
ENSDART00000134846
|
si:dkeyp-73g8.5
|
si:dkeyp-73g8.5 |
| chr14_-_2264494 | 0.27 |
ENSDART00000191149
|
pcdh2ab9
|
protocadherin 2 alpha b 9 |
| chr8_-_6877390 | 0.27 |
ENSDART00000170883
ENSDART00000005321 |
neflb
|
neurofilament, light polypeptide b |
| chr9_+_38292947 | 0.26 |
ENSDART00000146663
|
tfcp2l1
|
transcription factor CP2-like 1 |
| chr2_-_37889111 | 0.26 |
ENSDART00000168939
ENSDART00000098529 |
mbl2
|
mannose binding lectin 2 |
| chr4_+_74131530 | 0.26 |
ENSDART00000174125
|
CABZ01072523.1
|
|
| chr22_+_35516440 | 0.26 |
ENSDART00000184191
|
hykk.2
|
hydroxylysine kinase, tandem duplicate 2 |
| chr13_-_20381485 | 0.26 |
ENSDART00000131351
|
si:ch211-270n8.1
|
si:ch211-270n8.1 |
| chr16_-_26727032 | 0.26 |
ENSDART00000177668
|
rnf41l
|
ring finger protein 41, like |
| chr4_-_12388535 | 0.26 |
ENSDART00000017180
|
rergla
|
RERG/RAS-like a |
| chr17_-_25563847 | 0.25 |
ENSDART00000040032
|
rtn4ip1
|
reticulon 4 interacting protein 1 |
| chr7_-_59054322 | 0.25 |
ENSDART00000165390
|
chmp5b
|
charged multivesicular body protein 5b |
| chr1_-_999556 | 0.25 |
ENSDART00000170884
ENSDART00000172235 |
gart
|
phosphoribosylglycinamide formyltransferase |
| chr12_-_36260532 | 0.25 |
ENSDART00000022533
|
kcnj2a
|
potassium inwardly-rectifying channel, subfamily J, member 2a |
| chr4_+_61171310 | 0.25 |
ENSDART00000141738
|
si:dkey-9p20.18
|
si:dkey-9p20.18 |
| chr5_-_38179220 | 0.25 |
ENSDART00000147701
|
si:ch211-284e13.11
|
si:ch211-284e13.11 |
| chr11_-_17964525 | 0.24 |
ENSDART00000018948
|
cishb
|
cytokine inducible SH2-containing protein b |
| chr17_+_16873417 | 0.24 |
ENSDART00000146276
|
dio2
|
iodothyronine deiodinase 2 |
| chr19_-_12404590 | 0.24 |
ENSDART00000103703
|
ftr56
|
finTRIM family, member 56 |
| chr9_+_6587364 | 0.24 |
ENSDART00000122279
|
fhl2a
|
four and a half LIM domains 2a |
| chr4_+_43700319 | 0.24 |
ENSDART00000141967
|
si:ch211-226o13.1
|
si:ch211-226o13.1 |
| chr7_+_12835048 | 0.24 |
ENSDART00000016465
|
cx36.7
|
connexin 36.7 |
| chr6_-_15096556 | 0.23 |
ENSDART00000185327
|
fhl2b
|
four and a half LIM domains 2b |
| chr21_+_19858627 | 0.23 |
ENSDART00000147010
|
fybb
|
FYN binding protein b |
| chr9_+_13999620 | 0.23 |
ENSDART00000143229
|
cd28l
|
cd28-like molecule |
| chr13_+_50375800 | 0.22 |
ENSDART00000099537
|
cox5b2
|
cytochrome c oxidase subunit Vb 2 |
| chr17_-_39786222 | 0.22 |
ENSDART00000154515
|
pimr62
|
Pim proto-oncogene, serine/threonine kinase, related 62 |
| chr5_-_69041102 | 0.22 |
ENSDART00000161561
|
ank1a
|
ankyrin 1, erythrocytic a |
| chr9_+_16449398 | 0.22 |
ENSDART00000006787
|
epha3
|
eph receptor A3 |
| chr16_+_46430627 | 0.22 |
ENSDART00000127681
|
rpz6
|
rapunzel 6 |
| chr12_+_36109507 | 0.22 |
ENSDART00000175409
|
map2k6
|
mitogen-activated protein kinase kinase 6 |
| chr21_+_27448856 | 0.22 |
ENSDART00000100784
|
cfbl
|
complement factor b-like |
| chr4_-_8030583 | 0.22 |
ENSDART00000113628
|
si:ch211-240l19.8
|
si:ch211-240l19.8 |
| chr8_+_52642869 | 0.21 |
ENSDART00000163617
ENSDART00000189997 |
si:dkey-90l8.3
|
si:dkey-90l8.3 |
| chr18_+_17660158 | 0.21 |
ENSDART00000186279
|
cpne2
|
copine II |
| chr19_-_27858033 | 0.21 |
ENSDART00000103898
ENSDART00000144884 |
srd5a1
|
steroid-5-alpha-reductase, alpha polypeptide 1 (3-oxo-5 alpha-steroid delta 4-dehydrogenase alpha 1) |
| chr6_+_27452289 | 0.20 |
ENSDART00000186265
|
sned1
|
sushi, nidogen and EGF-like domains 1 |
| chr2_-_41861040 | 0.20 |
ENSDART00000045763
|
keap1a
|
kelch-like ECH-associated protein 1a |
| chr6_+_39905021 | 0.20 |
ENSDART00000064904
|
endou
|
endonuclease, polyU-specific |
| chr18_+_17428258 | 0.20 |
ENSDART00000010452
|
zgc:91860
|
zgc:91860 |
| chr4_-_38033800 | 0.20 |
ENSDART00000159662
|
si:dkeyp-82b4.4
|
si:dkeyp-82b4.4 |
| chr2_+_40294313 | 0.20 |
ENSDART00000037292
|
epha4b
|
eph receptor A4b |
| chr6_-_49078263 | 0.19 |
ENSDART00000032982
|
slc5a8l
|
solute carrier family 5 (iodide transporter), member 8-like |
| chr16_-_29452039 | 0.19 |
ENSDART00000148960
|
si:ch211-113g11.6
|
si:ch211-113g11.6 |
| chr2_-_37043540 | 0.18 |
ENSDART00000131834
|
gng7
|
guanine nucleotide binding protein (G protein), gamma 7 |
| chr17_-_45552602 | 0.18 |
ENSDART00000154844
ENSDART00000034432 |
susd4
|
sushi domain containing 4 |
| chr22_-_35330532 | 0.18 |
ENSDART00000172654
|
FO818743.1
|
|
| chr18_+_45542981 | 0.18 |
ENSDART00000140357
|
kifc3
|
kinesin family member C3 |
| chr19_-_7144548 | 0.17 |
ENSDART00000147177
ENSDART00000134850 |
psmb8a
psmb13a
|
proteasome subunit beta 8A proteasome subunit beta 13a |
| chr22_-_37611681 | 0.17 |
ENSDART00000028085
|
ttc14
|
tetratricopeptide repeat domain 14 |
| chr15_+_5321612 | 0.17 |
ENSDART00000174345
|
or113-1
|
odorant receptor, family A, subfamily 113, member 1 |
| chr14_+_28438947 | 0.17 |
ENSDART00000006489
|
acsl4a
|
acyl-CoA synthetase long chain family member 4a |
| chr7_-_26457208 | 0.16 |
ENSDART00000173519
|
zgc:172079
|
zgc:172079 |
| chr14_-_25444774 | 0.16 |
ENSDART00000183448
|
slc26a2
|
solute carrier family 26 (anion exchanger), member 2 |
| chr6_+_9870192 | 0.16 |
ENSDART00000150894
|
MPP4 (1 of many)
|
si:ch211-222n4.6 |
| chr18_-_21177674 | 0.15 |
ENSDART00000060175
|
si:dkey-12e7.4
|
si:dkey-12e7.4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nfixa+nfixb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0071831 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 0.5 | 1.4 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) |
| 0.4 | 1.5 | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling(GO:0010882) |
| 0.4 | 1.1 | GO:1902176 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902176) |
| 0.3 | 1.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.2 | 1.0 | GO:0010332 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.2 | 0.6 | GO:0097376 | interneuron axon guidance(GO:0097376) spinal cord interneuron axon guidance(GO:0097377) dorsal spinal cord interneuron axon guidance(GO:0097378) |
| 0.2 | 0.2 | GO:0002820 | negative regulation of adaptive immune response(GO:0002820) negative regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains(GO:0002823) |
| 0.2 | 1.5 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.2 | 0.6 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.1 | 0.4 | GO:1902895 | pri-miRNA transcription from RNA polymerase II promoter(GO:0061614) regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902893) positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.1 | 0.7 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.1 | 0.7 | GO:0033986 | response to methanol(GO:0033986) cellular response to methanol(GO:0071405) |
| 0.1 | 0.6 | GO:0006660 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.1 | 2.5 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 0.3 | GO:0002468 | dendritic cell antigen processing and presentation(GO:0002468) |
| 0.1 | 0.6 | GO:0035176 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.1 | 3.9 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 0.6 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 1.9 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.1 | 2.5 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 0.4 | GO:0042762 | regulation of sulfur metabolic process(GO:0042762) |
| 0.1 | 0.6 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.1 | 0.2 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.1 | 0.5 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 0.3 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 0.8 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.1 | 0.5 | GO:1901099 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.1 | 0.3 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.1 | 0.9 | GO:0036065 | fucosylation(GO:0036065) |
| 0.1 | 0.3 | GO:0046084 | adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.1 | 0.3 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 | 1.2 | GO:0072176 | pronephric duct development(GO:0039022) nephric duct development(GO:0072176) |
| 0.0 | 1.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.5 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.3 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.0 | 0.2 | GO:1900407 | regulation of cellular response to oxidative stress(GO:1900407) |
| 0.0 | 0.2 | GO:0042403 | thyroid hormone metabolic process(GO:0042403) |
| 0.0 | 0.7 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.0 | 0.7 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.1 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.0 | 0.3 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.8 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.2 | GO:1900744 | regulation of p38MAPK cascade(GO:1900744) |
| 0.0 | 0.2 | GO:0070571 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.0 | 1.5 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.3 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.6 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.0 | 0.2 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.0 | 0.1 | GO:0030859 | polarized epithelial cell differentiation(GO:0030859) |
| 0.0 | 1.6 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.3 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.0 | 0.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.0 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.7 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.1 | GO:0032689 | negative regulation of interferon-gamma production(GO:0032689) CD4-positive, alpha-beta T cell proliferation(GO:0035739) regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.0 | 0.4 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.0 | 0.5 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.3 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.7 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.3 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 1.2 | GO:1990266 | neutrophil migration(GO:1990266) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.2 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.2 | 1.5 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.2 | 2.5 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 1.0 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.1 | 0.3 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 0.3 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.6 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.8 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 0.6 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.2 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 2.4 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.7 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 2.2 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 1.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.9 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.9 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.3 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.3 | GO:0019814 | immunoglobulin complex(GO:0019814) |
| 0.0 | 0.1 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 0.2 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.1 | GO:0033010 | paranodal junction(GO:0033010) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.2 | 2.5 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.2 | 0.6 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 0.7 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.1 | 0.7 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.1 | 1.1 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 0.5 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 0.8 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 1.5 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 0.6 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.1 | 0.9 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.6 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.1 | 0.5 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 1.4 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 2.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 1.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.5 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.1 | 0.3 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.1 | 0.2 | GO:0033765 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) enone reductase activity(GO:0035671) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.1 | 0.9 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 1.0 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.6 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 0.6 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 1.1 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.4 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.7 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.3 | GO:0019202 | amino acid kinase activity(GO:0019202) |
| 0.0 | 0.3 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.6 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.1 | GO:0008397 | sterol 12-alpha-hydroxylase activity(GO:0008397) |
| 0.0 | 0.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.3 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.0 | 1.3 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.8 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.3 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.0 | 0.4 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 0.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 1.5 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 1.7 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 2.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.5 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.6 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.2 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.5 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.3 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.3 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 1.7 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.6 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 1.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.2 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.6 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.2 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 1.0 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.7 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.0 | 0.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.3 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.2 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |