Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
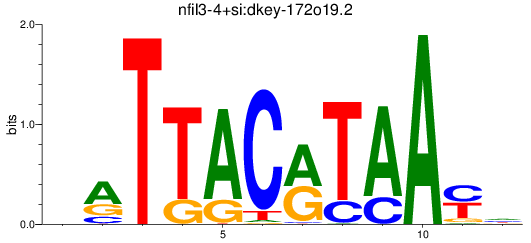
Results for nfil3-4+si:dkey-172o19.2
Z-value: 0.54
Transcription factors associated with nfil3-4+si:dkey-172o19.2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
si_dkey-172o19.2
|
ENSDARG00000071398 | si_dkey-172o19.2 |
|
nfil3-4
|
ENSDARG00000092346 | nuclear factor, interleukin 3 regulated, member 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| si:dkey-172o19.2 | dr11_v1_chr22_+_20546612_20546612 | -0.27 | 4.8e-01 | Click! |
| nfil3-4 | dr11_v1_chr22_+_20560041_20560041 | 0.00 | 1.0e+00 | Click! |
Activity profile of nfil3-4+si:dkey-172o19.2 motif
Sorted Z-values of nfil3-4+si:dkey-172o19.2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_12725513 | 0.63 |
ENSDART00000132286
|
mgst1.2
|
microsomal glutathione S-transferase 1.2 |
| chr13_+_502230 | 0.60 |
ENSDART00000013007
|
degs1
|
delta(4)-desaturase, sphingolipid 1 |
| chr23_-_23401305 | 0.56 |
ENSDART00000078936
|
her9
|
hairy-related 9 |
| chr25_-_29415369 | 0.56 |
ENSDART00000110774
ENSDART00000019183 |
ugt5a2
ugt5a1
|
UDP glucuronosyltransferase 5 family, polypeptide A2 UDP glucuronosyltransferase 5 family, polypeptide A1 |
| chr15_+_19838458 | 0.50 |
ENSDART00000101204
|
alcamb
|
activated leukocyte cell adhesion molecule b |
| chr20_-_35246150 | 0.48 |
ENSDART00000090549
|
fzd3a
|
frizzled class receptor 3a |
| chr7_-_24364536 | 0.48 |
ENSDART00000064789
|
txn
|
thioredoxin |
| chr5_+_6955900 | 0.47 |
ENSDART00000099417
|
CABZ01041962.1
|
|
| chr9_-_45602978 | 0.44 |
ENSDART00000139019
ENSDART00000085763 |
agr1
|
anterior gradient 1 |
| chr8_+_2487883 | 0.43 |
ENSDART00000101841
|
dynll1
|
dynein, light chain, LC8-type 1 |
| chr18_-_16922905 | 0.43 |
ENSDART00000187165
|
wee1
|
WEE1 G2 checkpoint kinase |
| chr25_+_24616717 | 0.42 |
ENSDART00000089113
|
abtb2b
|
ankyrin repeat and BTB (POZ) domain containing 2b |
| chr22_+_11775269 | 0.41 |
ENSDART00000140272
|
krt96
|
keratin 96 |
| chr19_+_11985572 | 0.41 |
ENSDART00000130537
|
spag1a
|
sperm associated antigen 1a |
| chr5_+_26121393 | 0.39 |
ENSDART00000002221
|
bco2l
|
beta-carotene 15, 15-dioxygenase 2, like |
| chr20_+_1272526 | 0.38 |
ENSDART00000008115
ENSDART00000133825 |
hsd3b2
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 |
| chr13_+_18533005 | 0.37 |
ENSDART00000136024
|
ftr14l
|
finTRIM family, member 14-like |
| chr5_+_37091626 | 0.36 |
ENSDART00000161054
|
tagln2
|
transgelin 2 |
| chr12_+_5708400 | 0.35 |
ENSDART00000017191
|
dlx3b
|
distal-less homeobox 3b |
| chr16_+_43139504 | 0.35 |
ENSDART00000065643
|
dbf4
|
DBF4 zinc finger |
| chr2_+_11685742 | 0.34 |
ENSDART00000138562
|
greb1l
|
growth regulation by estrogen in breast cancer-like |
| chr7_+_56577906 | 0.34 |
ENSDART00000184023
|
hp
|
haptoglobin |
| chr3_+_28594713 | 0.34 |
ENSDART00000179799
|
sept12
|
septin 12 |
| chr14_+_21107032 | 0.34 |
ENSDART00000138319
ENSDART00000139103 ENSDART00000184735 |
aldob
|
aldolase b, fructose-bisphosphate |
| chr9_-_48736388 | 0.34 |
ENSDART00000022074
|
dhrs9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr7_+_56577522 | 0.33 |
ENSDART00000149130
ENSDART00000149624 |
hp
|
haptoglobin |
| chr14_-_12071679 | 0.33 |
ENSDART00000165581
|
tmsb1
|
thymosin beta 1 |
| chr8_+_999421 | 0.32 |
ENSDART00000149528
|
fabp1b.1
|
fatty acid binding protein 1b, tandem duplicate 1 |
| chr14_+_21106444 | 0.32 |
ENSDART00000075744
ENSDART00000132363 |
aldob
|
aldolase b, fructose-bisphosphate |
| chr12_-_20584413 | 0.32 |
ENSDART00000170923
|
FP885542.2
|
|
| chr17_-_14876758 | 0.31 |
ENSDART00000155857
|
nid2a
|
nidogen 2a (osteonidogen) |
| chr5_-_65037525 | 0.31 |
ENSDART00000158856
|
anxa1b
|
annexin A1b |
| chr11_-_8167799 | 0.31 |
ENSDART00000133574
ENSDART00000024046 ENSDART00000146940 |
uox
|
urate oxidase |
| chr5_-_65037371 | 0.31 |
ENSDART00000170560
|
anxa1b
|
annexin A1b |
| chr21_-_37194365 | 0.30 |
ENSDART00000100286
|
fgfr4
|
fibroblast growth factor receptor 4 |
| chr10_-_20523405 | 0.30 |
ENSDART00000114824
|
ddhd2
|
DDHD domain containing 2 |
| chr14_-_12071447 | 0.30 |
ENSDART00000166116
|
tmsb1
|
thymosin beta 1 |
| chr1_-_35928942 | 0.30 |
ENSDART00000033566
|
smad1
|
SMAD family member 1 |
| chr4_-_77432218 | 0.30 |
ENSDART00000158683
|
slco1d1
|
solute carrier organic anion transporter family, member 1D1 |
| chr9_-_9225980 | 0.30 |
ENSDART00000180301
|
cbsb
|
cystathionine-beta-synthase b |
| chr12_+_31616412 | 0.30 |
ENSDART00000124439
|
cpn1
|
carboxypeptidase N, polypeptide 1 |
| chr3_-_10312232 | 0.29 |
ENSDART00000081766
|
nog1
|
noggin 1 |
| chr24_+_38671054 | 0.29 |
ENSDART00000154214
|
si:ch73-70c5.1
|
si:ch73-70c5.1 |
| chr20_-_30900947 | 0.29 |
ENSDART00000153419
ENSDART00000062536 |
hebp2
|
heme binding protein 2 |
| chr15_-_26636826 | 0.29 |
ENSDART00000087632
|
slc47a4
|
solute carrier family 47 (multidrug and toxin extrusion), member 4 |
| chr14_+_45406299 | 0.29 |
ENSDART00000173142
ENSDART00000112377 |
map1lc3cl
|
microtubule-associated protein 1 light chain 3 gamma, like |
| chr16_+_23984755 | 0.28 |
ENSDART00000145328
|
apoc2
|
apolipoprotein C-II |
| chr10_+_38526496 | 0.27 |
ENSDART00000144329
|
acer3
|
alkaline ceramidase 3 |
| chr10_+_2799285 | 0.27 |
ENSDART00000030709
|
pnx
|
posterior neuron-specific homeobox |
| chr4_-_2545310 | 0.26 |
ENSDART00000150619
ENSDART00000140760 |
e2f7
|
E2F transcription factor 7 |
| chr5_-_69212184 | 0.26 |
ENSDART00000053963
|
mat2ab
|
methionine adenosyltransferase II, alpha b |
| chr14_+_1170968 | 0.26 |
ENSDART00000125203
ENSDART00000193575 |
hopx
|
HOP homeobox |
| chr3_+_23092762 | 0.26 |
ENSDART00000142884
ENSDART00000024136 |
gngt2a
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2a |
| chr5_-_55560937 | 0.26 |
ENSDART00000148436
|
gna14
|
guanine nucleotide binding protein (G protein), alpha 14 |
| chr15_-_29388012 | 0.26 |
ENSDART00000115032
|
serpinh1b
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1b |
| chr21_+_25625026 | 0.26 |
ENSDART00000134678
|
ovol1b
|
ovo-like zinc finger 1b |
| chr15_-_29348212 | 0.26 |
ENSDART00000133117
|
tsku
|
tsukushi small leucine rich proteoglycan homolog (Xenopus laevis) |
| chr20_+_25563105 | 0.25 |
ENSDART00000063100
|
cyp2p6
|
cytochrome P450, family 2, subfamily P, polypeptide 6 |
| chr16_+_40576679 | 0.25 |
ENSDART00000169412
ENSDART00000193464 |
ccne2
|
cyclin E2 |
| chr2_+_47905735 | 0.25 |
ENSDART00000159701
|
ftr23
|
finTRIM family, member 23 |
| chr8_+_41647539 | 0.25 |
ENSDART00000136492
ENSDART00000138799 ENSDART00000134404 |
si:ch211-158d24.4
|
si:ch211-158d24.4 |
| chr16_-_51253925 | 0.25 |
ENSDART00000050644
|
serpinb14
|
serpin peptidase inhibitor, clade B (ovalbumin), member 14 |
| chr1_-_35929143 | 0.25 |
ENSDART00000185002
|
smad1
|
SMAD family member 1 |
| chr20_-_9760424 | 0.25 |
ENSDART00000104936
|
si:dkey-63j12.4
|
si:dkey-63j12.4 |
| chr16_-_30584279 | 0.24 |
ENSDART00000077226
|
mc5rb
|
melanocortin 5b receptor |
| chr9_-_456541 | 0.24 |
ENSDART00000159596
|
zgc:91849
|
zgc:91849 |
| chr5_+_61476014 | 0.24 |
ENSDART00000050906
|
lrwd1
|
leucine-rich repeats and WD repeat domain containing 1 |
| chr12_-_45197038 | 0.24 |
ENSDART00000016635
|
bccip
|
BRCA2 and CDKN1A interacting protein |
| chr5_+_61475451 | 0.24 |
ENSDART00000163444
|
lrwd1
|
leucine-rich repeats and WD repeat domain containing 1 |
| chr24_-_26310854 | 0.24 |
ENSDART00000080113
|
apodb
|
apolipoprotein Db |
| chr19_+_16015881 | 0.23 |
ENSDART00000187135
|
ctps1a
|
CTP synthase 1a |
| chr1_-_39895859 | 0.23 |
ENSDART00000135791
ENSDART00000035739 |
tmem134
|
transmembrane protein 134 |
| chr19_-_2231146 | 0.23 |
ENSDART00000181909
ENSDART00000043595 |
twist1a
|
twist family bHLH transcription factor 1a |
| chr4_+_30775376 | 0.23 |
ENSDART00000158528
|
si:dkey-11d20.1
|
si:dkey-11d20.1 |
| chr5_-_57655092 | 0.23 |
ENSDART00000074290
|
mia
|
melanoma inhibitory activity |
| chr22_+_13886821 | 0.23 |
ENSDART00000130585
ENSDART00000105711 |
sh3bp4a
|
SH3-domain binding protein 4a |
| chr18_+_16943911 | 0.23 |
ENSDART00000157609
|
si:dkey-8l13.5
|
si:dkey-8l13.5 |
| chr18_-_40905901 | 0.23 |
ENSDART00000064848
|
pglyrp5
|
peptidoglycan recognition protein 5 |
| chr5_+_25585869 | 0.23 |
ENSDART00000138060
|
si:dkey-229d2.7
|
si:dkey-229d2.7 |
| chr22_+_5478353 | 0.22 |
ENSDART00000160596
|
tppp
|
tubulin polymerization promoting protein |
| chr1_-_59411901 | 0.22 |
ENSDART00000167015
|
si:ch211-188p14.4
|
si:ch211-188p14.4 |
| chr22_-_11438627 | 0.22 |
ENSDART00000007649
|
mid1ip1b
|
MID1 interacting protein 1b |
| chr25_-_19585010 | 0.22 |
ENSDART00000021340
|
sycp3
|
synaptonemal complex protein 3 |
| chr9_-_33121535 | 0.22 |
ENSDART00000166371
ENSDART00000138052 |
zgc:172014
|
zgc:172014 |
| chr16_+_23984179 | 0.22 |
ENSDART00000175879
|
apoc2
|
apolipoprotein C-II |
| chr17_-_20236228 | 0.21 |
ENSDART00000136490
ENSDART00000029380 |
bnip4
|
BCL2 interacting protein 4 |
| chr15_+_618081 | 0.21 |
ENSDART00000181518
|
si:ch211-210b2.1
|
si:ch211-210b2.1 |
| chr22_-_15593824 | 0.21 |
ENSDART00000123125
|
tpm4a
|
tropomyosin 4a |
| chr12_+_27537357 | 0.21 |
ENSDART00000136212
|
etv4
|
ets variant 4 |
| chr4_-_43056627 | 0.21 |
ENSDART00000150269
|
si:dkey-54j5.2
|
si:dkey-54j5.2 |
| chr3_-_32071468 | 0.21 |
ENSDART00000121979
|
BX511080.1
|
|
| chr17_+_27456804 | 0.21 |
ENSDART00000017756
ENSDART00000181461 ENSDART00000180178 |
ctsl.1
|
cathepsin L.1 |
| chr3_-_32927516 | 0.21 |
ENSDART00000140117
|
aoc2
|
amine oxidase, copper containing 2 |
| chr2_+_26288301 | 0.21 |
ENSDART00000017668
|
ptbp1a
|
polypyrimidine tract binding protein 1a |
| chr17_+_26965351 | 0.21 |
ENSDART00000114215
ENSDART00000147192 |
grhl3
|
grainyhead-like transcription factor 3 |
| chr3_-_1204341 | 0.21 |
ENSDART00000089646
|
fam234b
|
family with sequence similarity 234, member B |
| chr20_-_27225876 | 0.21 |
ENSDART00000149204
ENSDART00000149732 |
si:dkey-85n7.7
|
si:dkey-85n7.7 |
| chr6_-_10752937 | 0.20 |
ENSDART00000135093
|
ola1
|
Obg-like ATPase 1 |
| chr15_+_15232855 | 0.20 |
ENSDART00000033904
ENSDART00000139263 |
med29
|
mediator complex subunit 29 |
| chr10_-_5016997 | 0.20 |
ENSDART00000101414
|
hnrnpd
|
heterogeneous nuclear ribonucleoprotein D |
| chr19_+_3653976 | 0.20 |
ENSDART00000125673
|
nedd9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| chr8_+_39634114 | 0.20 |
ENSDART00000144293
|
msi1
|
musashi RNA-binding protein 1 |
| chr6_+_50451337 | 0.20 |
ENSDART00000155051
|
mych
|
myelocytomatosis oncogene homolog |
| chr4_+_68088529 | 0.20 |
ENSDART00000159155
|
znf1096
|
zinc finger protein 1096 |
| chr13_-_32906265 | 0.20 |
ENSDART00000113823
ENSDART00000171114 ENSDART00000180035 ENSDART00000137570 |
si:dkey-18j18.3
|
si:dkey-18j18.3 |
| chr16_-_46619967 | 0.20 |
ENSDART00000158341
|
tmem176l.3a
|
transmembrane protein 176l.3a |
| chr25_+_34014523 | 0.20 |
ENSDART00000182856
|
anxa2a
|
annexin A2a |
| chr17_-_23412705 | 0.20 |
ENSDART00000126995
|
si:ch211-149k12.3
|
si:ch211-149k12.3 |
| chr5_-_20195350 | 0.20 |
ENSDART00000139675
|
dao.1
|
D-amino-acid oxidase, tandem duplicate 1 |
| chr14_-_28568107 | 0.20 |
ENSDART00000042850
ENSDART00000145502 |
insb
|
preproinsulin b |
| chr4_+_45504471 | 0.19 |
ENSDART00000150399
|
si:dkey-256i11.2
|
si:dkey-256i11.2 |
| chr16_-_29557338 | 0.19 |
ENSDART00000058888
|
hormad1
|
HORMA domain containing 1 |
| chr8_+_1187928 | 0.19 |
ENSDART00000127252
|
slc35d2
|
solute carrier family 35 (UDP-GlcNAc/UDP-glucose transporter), member D2 |
| chr20_+_25568694 | 0.19 |
ENSDART00000063107
ENSDART00000063128 |
cyp2p7
|
cytochrome P450, family 2, subfamily P, polypeptide 7 |
| chr17_+_30369396 | 0.19 |
ENSDART00000076611
|
greb1
|
growth regulation by estrogen in breast cancer 1 |
| chr11_+_6374448 | 0.19 |
ENSDART00000135906
|
AL840638.2
|
|
| chr6_-_27891961 | 0.19 |
ENSDART00000155116
|
im:7152348
|
im:7152348 |
| chr24_-_26328721 | 0.19 |
ENSDART00000125468
|
apodb
|
apolipoprotein Db |
| chr20_+_42668875 | 0.19 |
ENSDART00000048890
|
slc22a2
|
solute carrier family 22 (organic cation transporter), member 2 |
| chr25_+_35051656 | 0.19 |
ENSDART00000133379
|
hist2h3c
|
histone cluster 2, H3c |
| chr13_+_33688474 | 0.19 |
ENSDART00000161465
|
CABZ01087953.1
|
|
| chr6_+_16468776 | 0.19 |
ENSDART00000109151
ENSDART00000114667 |
zgc:161969
|
zgc:161969 |
| chr2_-_20923864 | 0.18 |
ENSDART00000006870
|
ptgs2a
|
prostaglandin-endoperoxide synthase 2a |
| chr6_-_41085692 | 0.18 |
ENSDART00000181463
|
srsf3a
|
serine/arginine-rich splicing factor 3a |
| chr7_-_27037990 | 0.18 |
ENSDART00000173561
|
nucb2a
|
nucleobindin 2a |
| chr24_-_17023392 | 0.18 |
ENSDART00000106058
|
ptgdsb.2
|
prostaglandin D2 synthase b, tandem duplicate 2 |
| chr24_-_8732519 | 0.18 |
ENSDART00000082351
|
tfap2a
|
transcription factor AP-2 alpha |
| chr15_-_47865063 | 0.18 |
ENSDART00000151600
|
h3f3b.1
|
H3 histone, family 3B.1 |
| chr7_-_12968689 | 0.18 |
ENSDART00000173115
ENSDART00000013690 |
rplp2l
|
ribosomal protein, large P2, like |
| chr19_+_15440841 | 0.18 |
ENSDART00000182329
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr24_+_4373355 | 0.18 |
ENSDART00000179062
ENSDART00000093256 ENSDART00000138943 |
ccny
|
cyclin Y |
| chr10_+_38417512 | 0.18 |
ENSDART00000112457
|
samsn1b
|
SAM domain, SH3 domain and nuclear localisation signals 1b |
| chr4_-_35900331 | 0.18 |
ENSDART00000164413
|
zgc:174653
|
zgc:174653 |
| chr3_-_16250527 | 0.18 |
ENSDART00000146699
ENSDART00000141181 |
bckdhbl
|
branched chain keto acid dehydrogenase E1, beta polypeptide, like |
| chr23_-_4925641 | 0.18 |
ENSDART00000140861
ENSDART00000060718 |
taz
|
tafazzin |
| chr10_+_33982010 | 0.18 |
ENSDART00000180431
|
fryb
|
furry homolog b (Drosophila) |
| chr4_+_64577406 | 0.18 |
ENSDART00000159754
|
si:ch211-223a21.4
|
si:ch211-223a21.4 |
| chr4_-_73739119 | 0.18 |
ENSDART00000108669
|
zgc:171551
|
zgc:171551 |
| chr25_-_19584735 | 0.18 |
ENSDART00000137930
|
sycp3
|
synaptonemal complex protein 3 |
| chr14_-_31087830 | 0.18 |
ENSDART00000002250
|
hs6st2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chr3_+_24458204 | 0.18 |
ENSDART00000155028
ENSDART00000153551 |
cbx6b
|
chromobox homolog 6b |
| chr3_-_38918697 | 0.18 |
ENSDART00000145630
|
si:dkey-106c17.2
|
si:dkey-106c17.2 |
| chr17_+_32374876 | 0.17 |
ENSDART00000183851
|
ywhaqb
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide b |
| chr6_-_24301324 | 0.17 |
ENSDART00000171401
|
BX927081.1
|
|
| chr23_+_27352072 | 0.17 |
ENSDART00000144762
|
si:dkey-9p24.5
|
si:dkey-9p24.5 |
| chr23_+_43489182 | 0.17 |
ENSDART00000162062
|
znf341
|
zinc finger protein 341 |
| chr22_-_21314821 | 0.17 |
ENSDART00000105546
ENSDART00000135388 |
cks2
|
CDC28 protein kinase regulatory subunit 2 |
| chr4_+_37952218 | 0.17 |
ENSDART00000186865
|
si:dkeyp-82b4.2
|
si:dkeyp-82b4.2 |
| chr4_+_58576146 | 0.17 |
ENSDART00000164911
|
si:ch211-212k5.4
|
si:ch211-212k5.4 |
| chr8_-_13013123 | 0.17 |
ENSDART00000147802
|
dennd2da
|
DENN/MADD domain containing 2Da |
| chr4_+_42556555 | 0.17 |
ENSDART00000168536
|
znf1053
|
zinc finger protein 1053 |
| chr13_+_24402406 | 0.17 |
ENSDART00000043002
|
rab1ab
|
RAB1A, member RAS oncogene family b |
| chr11_-_10770053 | 0.17 |
ENSDART00000179213
|
slc4a10a
|
solute carrier family 4, sodium bicarbonate transporter, member 10a |
| chr3_-_15734358 | 0.17 |
ENSDART00000137325
|
mvp
|
major vault protein |
| chr22_+_10606573 | 0.17 |
ENSDART00000192638
|
rad54l2
|
RAD54 like 2 |
| chr3_-_5394800 | 0.17 |
ENSDART00000181938
|
si:ch73-106l15.2
|
si:ch73-106l15.2 |
| chr4_+_39766931 | 0.17 |
ENSDART00000138663
|
si:dkeyp-85d8.1
|
si:dkeyp-85d8.1 |
| chr1_+_50538839 | 0.17 |
ENSDART00000020412
|
pkd2
|
polycystic kidney disease 2 |
| chr16_-_51299061 | 0.16 |
ENSDART00000148677
|
serpinb1l4
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1, like 4 |
| chr9_+_32301456 | 0.16 |
ENSDART00000078608
ENSDART00000185153 ENSDART00000144947 |
hspe1
|
heat shock 10 protein 1 |
| chr9_+_38158570 | 0.16 |
ENSDART00000059549
ENSDART00000133060 |
nifk
|
nucleolar protein interacting with the FHA domain of MKI67 |
| chr18_-_1228688 | 0.16 |
ENSDART00000064403
|
nptnb
|
neuroplastin b |
| chr24_+_119680 | 0.16 |
ENSDART00000061973
|
tgfbr1b
|
transforming growth factor, beta receptor 1 b |
| chr17_-_17764801 | 0.16 |
ENSDART00000155261
|
slirp
|
SRA stem-loop interacting RNA binding protein |
| chr6_+_50452699 | 0.16 |
ENSDART00000115049
|
mych
|
myelocytomatosis oncogene homolog |
| chr4_+_63818718 | 0.16 |
ENSDART00000161177
|
si:dkey-30f3.2
|
si:dkey-30f3.2 |
| chr16_+_23397785 | 0.16 |
ENSDART00000148961
|
s100a10b
|
S100 calcium binding protein A10b |
| chr24_+_16983269 | 0.16 |
ENSDART00000139176
ENSDART00000023833 |
eif2s3
|
eukaryotic translation initiation factor 2, subunit 3 gamma |
| chr5_-_36949476 | 0.16 |
ENSDART00000047269
|
h3f3c
|
H3 histone, family 3C |
| chr5_-_36948586 | 0.16 |
ENSDART00000193606
|
h3f3c
|
H3 histone, family 3C |
| chr4_+_14971239 | 0.16 |
ENSDART00000005985
|
smo
|
smoothened, frizzled class receptor |
| chr10_+_33895315 | 0.16 |
ENSDART00000142881
|
fryb
|
furry homolog b (Drosophila) |
| chr17_+_51744450 | 0.16 |
ENSDART00000190955
ENSDART00000149807 |
odc1
|
ornithine decarboxylase 1 |
| chr23_-_44546124 | 0.16 |
ENSDART00000126779
ENSDART00000024082 |
psmb6
|
proteasome subunit beta 6 |
| chr4_-_22671469 | 0.16 |
ENSDART00000050753
|
cd36
|
CD36 molecule (thrombospondin receptor) |
| chr25_+_30074947 | 0.16 |
ENSDART00000154849
|
si:ch211-147k10.5
|
si:ch211-147k10.5 |
| chr20_+_34326874 | 0.16 |
ENSDART00000061659
|
ivns1abpa
|
influenza virus NS1A binding protein a |
| chr5_+_15202495 | 0.16 |
ENSDART00000144915
|
tbx1
|
T-box 1 |
| chr20_-_3319642 | 0.16 |
ENSDART00000186743
ENSDART00000123096 |
marcksa
|
myristoylated alanine-rich protein kinase C substrate a |
| chr7_-_60831082 | 0.16 |
ENSDART00000073654
ENSDART00000136999 |
pcxb
|
pyruvate carboxylase b |
| chr19_+_16016038 | 0.16 |
ENSDART00000131319
|
ctps1a
|
CTP synthase 1a |
| chr25_-_18330503 | 0.16 |
ENSDART00000104496
|
dusp6
|
dual specificity phosphatase 6 |
| chr25_-_16600811 | 0.15 |
ENSDART00000011397
|
cpa2
|
carboxypeptidase A2 (pancreatic) |
| chr15_+_4969128 | 0.15 |
ENSDART00000062856
|
rnf169
|
ring finger protein 169 |
| chr21_+_22845317 | 0.15 |
ENSDART00000065555
|
birc2
|
baculoviral IAP repeat containing 2 |
| chr5_-_2112030 | 0.15 |
ENSDART00000091932
|
gusb
|
glucuronidase, beta |
| chr13_-_45942000 | 0.15 |
ENSDART00000158534
ENSDART00000171822 ENSDART00000166442 |
si:ch211-62a1.4
|
si:ch211-62a1.4 |
| chr9_+_5243476 | 0.15 |
ENSDART00000182735
|
CABZ01075242.1
|
|
| chr20_-_54245256 | 0.15 |
ENSDART00000170482
|
prpf39
|
PRP39 pre-mRNA processing factor 39 homolog (yeast) |
| chr25_+_34845469 | 0.15 |
ENSDART00000145416
|
tmem231
|
transmembrane protein 231 |
| chr4_-_45301719 | 0.15 |
ENSDART00000150282
|
si:ch211-162i8.2
|
si:ch211-162i8.2 |
| chr14_-_7786260 | 0.15 |
ENSDART00000128218
|
tmem173
|
transmembrane protein 173 |
| chr2_+_24762567 | 0.15 |
ENSDART00000078866
|
ifi30
|
interferon, gamma-inducible protein 30 |
| chr10_-_42923385 | 0.15 |
ENSDART00000076731
|
ACOT12
|
acyl-CoA thioesterase 12 |
| chr4_+_33461796 | 0.15 |
ENSDART00000150445
|
si:dkey-247i3.1
|
si:dkey-247i3.1 |
| chr24_+_39638555 | 0.15 |
ENSDART00000078313
|
luc7l
|
LUC7-like (S. cerevisiae) |
| chr16_+_23398369 | 0.15 |
ENSDART00000037694
|
s100a10b
|
S100 calcium binding protein A10b |
| chr3_-_50139860 | 0.15 |
ENSDART00000101563
|
btr02
|
bloodthirsty-related gene family, member 2 |
| chr17_+_1323699 | 0.15 |
ENSDART00000172540
|
adssl1
|
adenylosuccinate synthase like 1 |
| chr15_-_5780593 | 0.15 |
ENSDART00000163555
ENSDART00000082388 |
chchd2
|
coiled-coil-helix-coiled-coil-helix domain containing 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nfil3-4+si:dkey-172o19.2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902176) |
| 0.1 | 0.5 | GO:0071169 | establishment of protein localization to chromosome(GO:0070199) establishment of protein localization to chromatin(GO:0071169) |
| 0.1 | 0.4 | GO:0009912 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.1 | 0.6 | GO:0033034 | neutrophil apoptotic process(GO:0001781) regulation of T-helper 1 type immune response(GO:0002825) positive regulation of T-helper 1 type immune response(GO:0002827) negative regulation of type 2 immune response(GO:0002829) inflammatory cell apoptotic process(GO:0006925) regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of phospholipase A2 activity(GO:0032429) interleukin-2 production(GO:0032623) regulation of interleukin-2 production(GO:0032663) positive regulation of interleukin-2 production(GO:0032743) myeloid cell apoptotic process(GO:0033028) regulation of neutrophil apoptotic process(GO:0033029) positive regulation of neutrophil apoptotic process(GO:0033031) regulation of myeloid cell apoptotic process(GO:0033032) positive regulation of myeloid cell apoptotic process(GO:0033034) T-helper 1 type immune response(GO:0042088) positive regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043372) T-helper 1 cell differentiation(GO:0045063) positive regulation of T-helper cell differentiation(GO:0045624) regulation of T-helper 1 cell differentiation(GO:0045625) positive regulation of T-helper 1 cell differentiation(GO:0045627) regulation of T-helper 2 cell differentiation(GO:0045628) negative regulation of T-helper 2 cell differentiation(GO:0045629) positive regulation of fatty acid biosynthetic process(GO:0045723) positive regulation of alpha-beta T cell differentiation(GO:0046638) neutrophil clearance(GO:0097350) negative regulation of phospholipase A2 activity(GO:1900138) positive regulation of CD4-positive, alpha-beta T cell activation(GO:2000516) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.1 | 0.3 | GO:0046415 | urate catabolic process(GO:0019628) urate metabolic process(GO:0046415) |
| 0.1 | 0.3 | GO:0070857 | regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.1 | 0.3 | GO:0031335 | regulation of sulfur amino acid metabolic process(GO:0031335) regulation of homocysteine metabolic process(GO:0050666) |
| 0.1 | 0.4 | GO:0046144 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.1 | 0.6 | GO:2000105 | positive regulation of DNA-dependent DNA replication(GO:2000105) |
| 0.1 | 0.5 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.2 | GO:0007585 | respiratory gaseous exchange(GO:0007585) regulation of respiratory gaseous exchange(GO:0043576) |
| 0.1 | 0.8 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.1 | 0.5 | GO:0021885 | forebrain cell migration(GO:0021885) |
| 0.1 | 0.1 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 0.3 | GO:0003418 | chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 0.1 | 0.2 | GO:0060844 | arterial endothelial cell fate commitment(GO:0060844) |
| 0.1 | 0.2 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.1 | 0.2 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.1 | 0.2 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.1 | 0.2 | GO:0071498 | cellular response to fluid shear stress(GO:0071498) |
| 0.1 | 0.2 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.1 | 0.3 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.1 | 0.2 | GO:0003156 | regulation of organ formation(GO:0003156) |
| 0.1 | 0.2 | GO:0060468 | negative regulation of fertilization(GO:0060467) prevention of polyspermy(GO:0060468) cortical granule exocytosis(GO:0060471) |
| 0.0 | 0.2 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.0 | 0.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.1 | GO:2000434 | regulation of protein neddylation(GO:2000434) |
| 0.0 | 0.2 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.2 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.0 | 0.2 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.2 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.0 | 0.2 | GO:0060547 | negative regulation of necroptotic process(GO:0060546) negative regulation of necrotic cell death(GO:0060547) |
| 0.0 | 0.3 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.5 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.0 | 0.1 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.1 | GO:0090220 | telomere localization(GO:0034397) telomere tethering at nuclear periphery(GO:0034398) meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic telomere clustering(GO:0045141) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.1 | GO:1903504 | regulation of spindle checkpoint(GO:0090231) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.7 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.2 | GO:1902369 | negative regulation of RNA catabolic process(GO:1902369) |
| 0.0 | 0.2 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.1 | GO:1901296 | negative regulation of striated muscle cell differentiation(GO:0051154) negative regulation of cardiac muscle tissue development(GO:0055026) cardiac muscle cell fate commitment(GO:0060923) canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) negative regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901296) negative regulation of cardiocyte differentiation(GO:1905208) negative regulation of cardiac muscle cell differentiation(GO:2000726) |
| 0.0 | 0.1 | GO:2000815 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.0 | 0.1 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.3 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.3 | GO:0042664 | negative regulation of endodermal cell fate specification(GO:0042664) |
| 0.0 | 0.1 | GO:1903292 | protein localization to Golgi membrane(GO:1903292) |
| 0.0 | 0.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.1 | GO:0032207 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) |
| 0.0 | 0.1 | GO:0046689 | response to mercury ion(GO:0046689) detoxification of mercury ion(GO:0050787) |
| 0.0 | 0.3 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.0 | 0.1 | GO:0015865 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.0 | 0.6 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.1 | GO:1901890 | positive regulation of cell junction assembly(GO:1901890) |
| 0.0 | 0.1 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.1 | GO:0042364 | water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.0 | 0.1 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 0.2 | GO:0060420 | regulation of heart growth(GO:0060420) |
| 0.0 | 0.0 | GO:2000108 | positive regulation of leukocyte apoptotic process(GO:2000108) |
| 0.0 | 0.3 | GO:0045823 | positive regulation of heart contraction(GO:0045823) |
| 0.0 | 0.2 | GO:0016137 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.0 | 0.4 | GO:0039014 | pronephric nephron tubule epithelial cell differentiation(GO:0035778) cell differentiation involved in pronephros development(GO:0039014) |
| 0.0 | 0.1 | GO:0034088 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.0 | 0.1 | GO:0051454 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.0 | 0.1 | GO:0006083 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.0 | 0.1 | GO:0019344 | homoserine metabolic process(GO:0009092) cysteine biosynthetic process via cystathionine(GO:0019343) cysteine biosynthetic process(GO:0019344) transsulfuration(GO:0019346) |
| 0.0 | 0.1 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.0 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.0 | 0.1 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.5 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.0 | 0.1 | GO:0018904 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.0 | 0.1 | GO:0060307 | regulation of membrane repolarization(GO:0060306) regulation of ventricular cardiac muscle cell membrane repolarization(GO:0060307) regulation of cardiac muscle cell membrane repolarization(GO:0099623) ventricular cardiac muscle cell membrane repolarization(GO:0099625) |
| 0.0 | 0.2 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.1 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.0 | 0.1 | GO:0051230 | mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.0 | 0.2 | GO:0032099 | negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.0 | 0.3 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.0 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.0 | 0.3 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.3 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:0006596 | polyamine biosynthetic process(GO:0006596) |
| 0.0 | 0.3 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.1 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.0 | 0.3 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.0 | GO:0045579 | positive regulation of B cell differentiation(GO:0045579) |
| 0.0 | 0.1 | GO:0006562 | proline catabolic process(GO:0006562) |
| 0.0 | 0.0 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to ciliary membrane(GO:1903441) |
| 0.0 | 0.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.0 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.4 | GO:0007568 | aging(GO:0007568) |
| 0.0 | 0.1 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.0 | 0.0 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) |
| 0.0 | 0.1 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.1 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.1 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 | 0.0 | GO:0046824 | regulation of nucleobase-containing compound transport(GO:0032239) positive regulation of nucleobase-containing compound transport(GO:0032241) positive regulation of nucleocytoplasmic transport(GO:0046824) regulation of RNA export from nucleus(GO:0046831) positive regulation of RNA export from nucleus(GO:0046833) |
| 0.0 | 0.1 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.0 | 0.1 | GO:0045901 | positive regulation of translational elongation(GO:0045901) |
| 0.0 | 0.4 | GO:0007286 | spermatid development(GO:0007286) spermatid differentiation(GO:0048515) |
| 0.0 | 0.0 | GO:0097435 | fibril organization(GO:0097435) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.1 | GO:0010884 | positive regulation of lipid storage(GO:0010884) |
| 0.0 | 0.0 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.0 | 0.1 | GO:0034695 | response to prostaglandin E(GO:0034695) cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.0 | 0.1 | GO:0036372 | opsin transport(GO:0036372) |
| 0.0 | 0.2 | GO:0060324 | face development(GO:0060324) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.1 | GO:0030431 | sleep(GO:0030431) |
| 0.0 | 0.1 | GO:0052651 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.2 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 0.0 | GO:0003250 | cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) endocardial cell development(GO:0060958) cell proliferation involved in heart valve development(GO:2000793) |
| 0.0 | 0.0 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.1 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.2 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.3 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 1.2 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 0.0 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.0 | GO:0010633 | negative regulation of epithelial cell migration(GO:0010633) |
| 0.0 | 0.0 | GO:0010898 | regulation of triglyceride catabolic process(GO:0010896) positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.0 | 0.1 | GO:0031106 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.0 | 0.0 | GO:1901881 | positive regulation of protein depolymerization(GO:1901881) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 0.2 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.1 | 0.2 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.0 | 0.5 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.1 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.0 | 0.7 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.5 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.1 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.2 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.2 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.1 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.5 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.1 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.1 | GO:0005880 | nuclear microtubule(GO:0005880) |
| 0.0 | 0.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.4 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.1 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.3 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.1 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.0 | 0.1 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.0 | GO:0000941 | condensed nuclear chromosome inner kinetochore(GO:0000941) |
| 0.0 | 0.4 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.0 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.1 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.2 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.1 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 0.0 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.0 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.0 | 0.2 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.2 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.2 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 0.8 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.4 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.1 | 0.3 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 0.4 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 0.3 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.1 | 0.3 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.1 | 0.2 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.1 | 0.2 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 0.3 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.1 | 0.2 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.1 | 0.2 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.0 | 0.1 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.2 | GO:0005461 | UDP-glucuronic acid transmembrane transporter activity(GO:0005461) UDP-N-acetylgalactosamine transmembrane transporter activity(GO:0005463) |
| 0.0 | 0.2 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.3 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.3 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.2 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.3 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.2 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.0 | 0.2 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.3 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.0 | 0.5 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.3 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.0 | 0.5 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.2 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.0 | 0.2 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.0 | 0.1 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.2 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.2 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.2 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.0 | 0.1 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.0 | 0.5 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.6 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.3 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.1 | GO:0030273 | melanin-concentrating hormone receptor activity(GO:0030273) |
| 0.0 | 0.1 | GO:0050218 | propionate-CoA ligase activity(GO:0050218) |
| 0.0 | 0.4 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.1 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.0 | 0.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.2 | GO:0004586 | ornithine decarboxylase activity(GO:0004586) |
| 0.0 | 0.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.5 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.5 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.2 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.2 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.0 | 0.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.4 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.0 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.2 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.0 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.0 | 0.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.2 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.0 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.0 | 0.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.2 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.2 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.0 | GO:0051499 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.0 | 0.0 | GO:0001734 | mRNA (N6-adenosine)-methyltransferase activity(GO:0001734) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.5 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.1 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.1 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 0.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 0.5 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.4 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.3 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.4 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.3 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.1 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.2 | REACTOME EARLY PHASE OF HIV LIFE CYCLE | Genes involved in Early Phase of HIV Life Cycle |
| 0.0 | 0.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.1 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.2 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.3 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.4 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 0.3 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.1 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.2 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.1 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |