Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
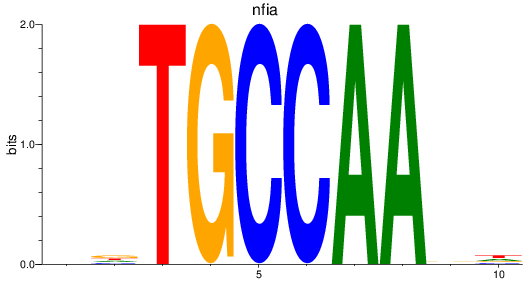
Results for nfia
Z-value: 2.43
Transcription factors associated with nfia
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nfia
|
ENSDARG00000062420 | nuclear factor I/A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nfia | dr11_v1_chr22_-_16997887_16997887 | 0.68 | 4.6e-02 | Click! |
Activity profile of nfia motif
Sorted Z-values of nfia motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_55139127 | 6.12 |
ENSDART00000115324
|
hbae1.3
|
hemoglobin, alpha embryonic 1.3 |
| chr25_+_20089986 | 4.09 |
ENSDART00000143441
ENSDART00000184073 |
tnni4b.2
|
troponin I4b, tandem duplicate 2 |
| chr12_-_17712393 | 3.80 |
ENSDART00000143534
ENSDART00000010144 |
pvalb2
|
parvalbumin 2 |
| chr5_-_32292965 | 3.78 |
ENSDART00000183522
ENSDART00000131983 |
myhz1.2
|
myosin, heavy polypeptide 1.2, skeletal muscle |
| chr3_+_23221047 | 3.78 |
ENSDART00000009393
|
col1a1a
|
collagen, type I, alpha 1a |
| chr21_+_26390549 | 3.55 |
ENSDART00000185643
|
tmsb
|
thymosin, beta |
| chr12_+_42436328 | 3.51 |
ENSDART00000167324
|
ebf3a
|
early B cell factor 3a |
| chr13_+_30804367 | 3.38 |
ENSDART00000053946
|
cxcl12a
|
chemokine (C-X-C motif) ligand 12a (stromal cell-derived factor 1) |
| chr16_-_55028740 | 3.35 |
ENSDART00000156368
ENSDART00000161704 |
zgc:114181
|
zgc:114181 |
| chr9_-_22182396 | 3.08 |
ENSDART00000101809
|
crygm2d6
|
crystallin, gamma M2d6 |
| chr5_+_32228538 | 3.07 |
ENSDART00000077471
|
myhc4
|
myosin heavy chain 4 |
| chr2_+_2470687 | 3.06 |
ENSDART00000184024
ENSDART00000061955 |
myl13
|
myosin, light chain 13 |
| chr7_+_29951997 | 3.04 |
ENSDART00000173453
|
tpma
|
alpha-tropomyosin |
| chr5_-_71705191 | 3.03 |
ENSDART00000187767
|
ak1
|
adenylate kinase 1 |
| chr15_-_23645810 | 2.94 |
ENSDART00000168845
|
ckmb
|
creatine kinase, muscle b |
| chr24_+_2519761 | 2.92 |
ENSDART00000106619
|
nrn1a
|
neuritin 1a |
| chr9_-_42696408 | 2.87 |
ENSDART00000144744
|
col5a2a
|
collagen, type V, alpha 2a |
| chr5_-_32309129 | 2.80 |
ENSDART00000123003
|
myhz1.1
|
myosin, heavy polypeptide 1.1, skeletal muscle |
| chr12_+_13256415 | 2.72 |
ENSDART00000144542
|
atp2a1l
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1, like |
| chr16_-_13595027 | 2.72 |
ENSDART00000060004
|
ntd5
|
ntl-dependent gene 5 |
| chr3_+_31621774 | 2.71 |
ENSDART00000076636
|
fzd2
|
frizzled class receptor 2 |
| chr11_+_30729745 | 2.64 |
ENSDART00000103270
|
slc22a7a
|
solute carrier family 22 (organic anion transporter), member 7a |
| chr5_+_32222303 | 2.63 |
ENSDART00000051362
|
myhc4
|
myosin heavy chain 4 |
| chr5_+_32247310 | 2.62 |
ENSDART00000182649
|
myha
|
myosin, heavy chain a |
| chr9_-_33107237 | 2.62 |
ENSDART00000013918
|
casq2
|
calsequestrin 2 |
| chr22_+_38767780 | 2.50 |
ENSDART00000149499
|
alpi.1
|
alkaline phosphatase, intestinal, tandem duplicate 1 |
| chr6_+_13742899 | 2.45 |
ENSDART00000104722
|
cdk5r2a
|
cyclin-dependent kinase 5, regulatory subunit 2a (p39) |
| chr15_-_29388012 | 2.42 |
ENSDART00000115032
|
serpinh1b
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1b |
| chr23_+_20110086 | 2.38 |
ENSDART00000054664
|
tnnc1b
|
troponin C type 1b (slow) |
| chr16_+_10318893 | 2.37 |
ENSDART00000055380
|
tubb5
|
tubulin, beta 5 |
| chr19_+_12915498 | 2.37 |
ENSDART00000132892
|
cthrc1a
|
collagen triple helix repeat containing 1a |
| chr24_-_24038800 | 2.36 |
ENSDART00000080549
|
lyz
|
lysozyme |
| chr23_+_36653376 | 2.22 |
ENSDART00000053189
|
gpr182
|
G protein-coupled receptor 182 |
| chr18_-_23874929 | 2.20 |
ENSDART00000134910
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr16_+_29650698 | 2.20 |
ENSDART00000137153
|
tmod4
|
tropomodulin 4 (muscle) |
| chr22_+_5176255 | 2.15 |
ENSDART00000092647
|
cers1
|
ceramide synthase 1 |
| chr16_-_45209684 | 2.14 |
ENSDART00000184595
|
fxyd1
|
FXYD domain containing ion transport regulator 1 (phospholemman) |
| chr15_-_26552393 | 2.14 |
ENSDART00000150152
|
serpinf2b
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2b |
| chr20_+_25340814 | 2.12 |
ENSDART00000063028
|
ctgfa
|
connective tissue growth factor a |
| chr3_-_50865079 | 2.09 |
ENSDART00000164295
|
pmp22a
|
peripheral myelin protein 22a |
| chr15_-_29387446 | 2.08 |
ENSDART00000145976
ENSDART00000035096 |
serpinh1b
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1b |
| chr20_+_26702377 | 2.07 |
ENSDART00000077753
|
foxc1b
|
forkhead box C1b |
| chr12_+_42436920 | 2.06 |
ENSDART00000177303
|
ebf3a
|
early B cell factor 3a |
| chr23_+_31107685 | 2.05 |
ENSDART00000103448
|
tbx18
|
T-box 18 |
| chr25_+_34576067 | 2.05 |
ENSDART00000157519
|
trpm1b
|
transient receptor potential cation channel, subfamily M, member 1b |
| chr7_-_60831082 | 1.94 |
ENSDART00000073654
ENSDART00000136999 |
pcxb
|
pyruvate carboxylase b |
| chr20_-_20533865 | 1.94 |
ENSDART00000125039
|
six6b
|
SIX homeobox 6b |
| chr20_-_44496245 | 1.92 |
ENSDART00000012229
|
fkbp1b
|
FK506 binding protein 1b |
| chr17_+_33719415 | 1.90 |
ENSDART00000132294
|
fut8a
|
fucosyltransferase 8a (alpha (1,6) fucosyltransferase) |
| chr18_-_40481028 | 1.89 |
ENSDART00000134177
|
zgc:101040
|
zgc:101040 |
| chr13_-_29424454 | 1.89 |
ENSDART00000026765
|
slc18a3a
|
solute carrier family 18 (vesicular acetylcholine transporter), member 3a |
| chr10_-_26738209 | 1.88 |
ENSDART00000188590
|
fgf13b
|
fibroblast growth factor 13b |
| chr15_+_7187228 | 1.88 |
ENSDART00000109394
|
her13
|
hairy-related 13 |
| chr6_-_55958705 | 1.87 |
ENSDART00000155963
|
eya2
|
EYA transcriptional coactivator and phosphatase 2 |
| chr22_+_28446365 | 1.87 |
ENSDART00000189359
|
abi3bpb
|
ABI family, member 3 (NESH) binding protein b |
| chr15_+_19652807 | 1.86 |
ENSDART00000134321
ENSDART00000054426 |
lim2.3
|
lens intrinsic membrane protein 2.3 |
| chr11_-_7261717 | 1.81 |
ENSDART00000128959
|
zgc:113223
|
zgc:113223 |
| chr21_-_44104600 | 1.74 |
ENSDART00000044599
|
oatx
|
organic anion transporter X |
| chr9_-_6927587 | 1.71 |
ENSDART00000059092
|
tmem182a
|
transmembrane protein 182a |
| chr18_+_22994113 | 1.71 |
ENSDART00000171871
|
cbfb
|
core-binding factor, beta subunit |
| chr8_-_16259063 | 1.71 |
ENSDART00000057590
|
dmrta2
|
DMRT-like family A2 |
| chr25_+_14507567 | 1.69 |
ENSDART00000015681
|
dbx1b
|
developing brain homeobox 1b |
| chr17_+_10242166 | 1.68 |
ENSDART00000170420
|
clec14a
|
C-type lectin domain containing 14A |
| chr18_+_15706160 | 1.67 |
ENSDART00000131524
|
si:ch211-264e16.1
|
si:ch211-264e16.1 |
| chr23_+_21663631 | 1.67 |
ENSDART00000066125
|
dhrs3a
|
dehydrogenase/reductase (SDR family) member 3a |
| chr23_-_10175898 | 1.63 |
ENSDART00000146185
|
krt5
|
keratin 5 |
| chr5_-_28679135 | 1.63 |
ENSDART00000193585
|
tnc
|
tenascin C |
| chr14_-_21064199 | 1.62 |
ENSDART00000172099
|
si:dkey-74k8.3
|
si:dkey-74k8.3 |
| chr2_+_38161318 | 1.61 |
ENSDART00000044264
|
mmp14b
|
matrix metallopeptidase 14b (membrane-inserted) |
| chr21_+_21374277 | 1.60 |
ENSDART00000079431
|
rtn2b
|
reticulon 2b |
| chr11_-_25418856 | 1.60 |
ENSDART00000013714
|
gata1a
|
GATA binding protein 1a |
| chr17_-_21793113 | 1.59 |
ENSDART00000104612
|
hmx3a
|
H6 family homeobox 3a |
| chr4_-_16412084 | 1.57 |
ENSDART00000188460
|
dcn
|
decorin |
| chr10_+_39084354 | 1.56 |
ENSDART00000158245
|
si:ch73-1a9.3
|
si:ch73-1a9.3 |
| chr16_+_20915319 | 1.56 |
ENSDART00000079383
|
hoxa9b
|
homeobox A9b |
| chr18_-_20636549 | 1.55 |
ENSDART00000060259
|
wnt2
|
wingless-type MMTV integration site family member 2 |
| chr4_+_19535946 | 1.54 |
ENSDART00000192342
ENSDART00000183740 ENSDART00000180812 ENSDART00000180017 |
lrrc4.1
|
leucine rich repeat containing 4.1 |
| chr7_-_17028015 | 1.53 |
ENSDART00000022441
|
dbx1a
|
developing brain homeobox 1a |
| chr14_-_21063977 | 1.53 |
ENSDART00000164373
|
si:dkey-74k8.3
|
si:dkey-74k8.3 |
| chr22_-_26945493 | 1.53 |
ENSDART00000077411
|
cxcl12b
|
chemokine (C-X-C motif) ligand 12b (stromal cell-derived factor 1) |
| chr22_+_5106751 | 1.53 |
ENSDART00000138967
|
atcaya
|
ataxia, cerebellar, Cayman type a |
| chr17_+_33453689 | 1.52 |
ENSDART00000156894
|
rin3
|
Ras and Rab interactor 3 |
| chr14_+_18785727 | 1.52 |
ENSDART00000184452
|
si:ch211-111e20.1
|
si:ch211-111e20.1 |
| chr14_-_32016615 | 1.51 |
ENSDART00000105761
|
zic3
|
zic family member 3 heterotaxy 1 (odd-paired homolog, Drosophila) |
| chr1_+_26667872 | 1.50 |
ENSDART00000152803
ENSDART00000152144 ENSDART00000152785 ENSDART00000152393 |
hemgn
|
hemogen |
| chr18_+_27318589 | 1.50 |
ENSDART00000037813
|
cd81b
|
CD81 molecule b |
| chr21_-_20932603 | 1.49 |
ENSDART00000138155
ENSDART00000079709 |
c6
|
complement component 6 |
| chr25_+_34014523 | 1.48 |
ENSDART00000182856
|
anxa2a
|
annexin A2a |
| chr16_-_38553721 | 1.47 |
ENSDART00000109124
|
rspo2
|
R-spondin 2 |
| chr17_+_18117358 | 1.47 |
ENSDART00000144894
|
bcl11ba
|
B cell CLL/lymphoma 11Ba |
| chr19_-_3381422 | 1.46 |
ENSDART00000105146
|
edn1
|
endothelin 1 |
| chr4_-_16341801 | 1.45 |
ENSDART00000140190
|
kera
|
keratocan |
| chr23_-_11870962 | 1.45 |
ENSDART00000143481
|
si:dkey-178k16.1
|
si:dkey-178k16.1 |
| chr3_-_53533128 | 1.44 |
ENSDART00000183591
|
notch3
|
notch 3 |
| chr22_+_5176693 | 1.44 |
ENSDART00000160927
|
cers1
|
ceramide synthase 1 |
| chr15_-_26552652 | 1.42 |
ENSDART00000152336
|
serpinf2b
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2b |
| chr19_-_9711472 | 1.41 |
ENSDART00000016197
ENSDART00000175075 |
slc2a3a
|
solute carrier family 2 (facilitated glucose transporter), member 3a |
| chr9_+_34089156 | 1.39 |
ENSDART00000000005
|
ccdc80
|
coiled-coil domain containing 80 |
| chr22_-_23253252 | 1.36 |
ENSDART00000175556
|
lhx9
|
LIM homeobox 9 |
| chr17_-_6382392 | 1.35 |
ENSDART00000188051
ENSDART00000192560 ENSDART00000137389 ENSDART00000115389 |
txlnbb
|
taxilin beta b |
| chr11_-_45185792 | 1.35 |
ENSDART00000171328
|
si:dkey-93h22.7
|
si:dkey-93h22.7 |
| chr7_+_18364176 | 1.35 |
ENSDART00000171606
ENSDART00000186368 |
cd248a
|
CD248 molecule, endosialin a |
| chr6_-_40771813 | 1.35 |
ENSDART00000076200
|
h1fx
|
H1 histone family, member X |
| chr10_-_17988779 | 1.34 |
ENSDART00000132206
ENSDART00000144841 |
si:dkey-242g16.2
|
si:dkey-242g16.2 |
| chr8_+_46217861 | 1.34 |
ENSDART00000038790
|
angptl7
|
angiopoietin-like 7 |
| chr10_-_28761454 | 1.34 |
ENSDART00000129400
|
alcama
|
activated leukocyte cell adhesion molecule a |
| chr17_+_51627209 | 1.33 |
ENSDART00000056886
|
zgc:113142
|
zgc:113142 |
| chr16_-_29277164 | 1.32 |
ENSDART00000058870
|
rhbg
|
Rh family, B glycoprotein (gene/pseudogene) |
| chr7_-_51321126 | 1.32 |
ENSDART00000067647
|
rasl11a
|
RAS-like, family 11, member A |
| chr19_+_19762183 | 1.30 |
ENSDART00000163611
ENSDART00000187604 |
hoxa3a
|
homeobox A3a |
| chr14_+_13454840 | 1.30 |
ENSDART00000161854
|
pls3
|
plastin 3 (T isoform) |
| chr16_+_31802203 | 1.30 |
ENSDART00000058739
ENSDART00000110834 |
wnt4b
|
wingless-type MMTV integration site family, member 4b |
| chr14_+_32022272 | 1.30 |
ENSDART00000105760
|
zic6
|
zic family member 6 |
| chr13_+_24834199 | 1.29 |
ENSDART00000101274
|
zgc:153981
|
zgc:153981 |
| chr23_+_23658474 | 1.29 |
ENSDART00000162838
|
agrn
|
agrin |
| chr10_-_22057001 | 1.29 |
ENSDART00000016575
|
tlx3b
|
T cell leukemia homeobox 3b |
| chr4_+_4232562 | 1.28 |
ENSDART00000177529
|
smkr1
|
small lysine rich protein 1 |
| chr3_+_32557615 | 1.27 |
ENSDART00000151608
|
pax10
|
paired box 10 |
| chr21_-_36972127 | 1.26 |
ENSDART00000100310
|
dbn1
|
drebrin 1 |
| chr5_-_51830997 | 1.26 |
ENSDART00000163616
|
homer1b
|
homer scaffolding protein 1b |
| chr15_+_27364394 | 1.26 |
ENSDART00000122101
|
tbx2b
|
T-box 2b |
| chr13_+_27951688 | 1.26 |
ENSDART00000050303
|
b3gat2
|
beta-1,3-glucuronyltransferase 2 (glucuronosyltransferase S) |
| chr25_-_15045338 | 1.25 |
ENSDART00000161165
ENSDART00000165774 ENSDART00000172538 |
pax6a
|
paired box 6a |
| chr15_-_34567370 | 1.25 |
ENSDART00000099793
|
sostdc1a
|
sclerostin domain containing 1a |
| chr5_-_41831646 | 1.24 |
ENSDART00000134326
|
si:dkey-65b12.6
|
si:dkey-65b12.6 |
| chr10_+_39263827 | 1.23 |
ENSDART00000172509
|
foxred1
|
FAD-dependent oxidoreductase domain containing 1 |
| chr18_+_44649804 | 1.23 |
ENSDART00000059063
|
ehd2b
|
EH-domain containing 2b |
| chr1_-_22861348 | 1.23 |
ENSDART00000139412
|
SMIM18
|
si:dkey-92j12.6 |
| chr25_+_28772632 | 1.23 |
ENSDART00000160619
|
slc41a2a
|
solute carrier family 41 (magnesium transporter), member 2a |
| chr5_+_37837245 | 1.22 |
ENSDART00000171617
|
epd
|
ependymin |
| chr11_+_13630107 | 1.22 |
ENSDART00000172220
|
si:ch211-1a19.3
|
si:ch211-1a19.3 |
| chr1_+_49568335 | 1.22 |
ENSDART00000142957
|
col17a1a
|
collagen, type XVII, alpha 1a |
| chr5_-_33460959 | 1.20 |
ENSDART00000085636
|
si:ch211-182d3.1
|
si:ch211-182d3.1 |
| chr14_-_30808174 | 1.20 |
ENSDART00000173262
|
prss23
|
protease, serine, 23 |
| chr17_-_19535328 | 1.20 |
ENSDART00000077809
|
cyp26c1
|
cytochrome P450, family 26, subfamily C, polypeptide 1 |
| chr5_-_67911111 | 1.19 |
ENSDART00000051833
|
gsx1
|
GS homeobox 1 |
| chr3_+_26044969 | 1.19 |
ENSDART00000133523
|
hmgxb4a
|
HMG box domain containing 4a |
| chr5_-_48268049 | 1.18 |
ENSDART00000187454
|
mef2cb
|
myocyte enhancer factor 2cb |
| chr21_+_11503212 | 1.17 |
ENSDART00000146701
|
si:dkey-184p9.7
|
si:dkey-184p9.7 |
| chr20_+_19066858 | 1.17 |
ENSDART00000192086
|
sox7
|
SRY (sex determining region Y)-box 7 |
| chr10_+_43189325 | 1.17 |
ENSDART00000185584
|
vcanb
|
versican b |
| chr15_-_20468302 | 1.17 |
ENSDART00000018514
|
dlc
|
deltaC |
| chr24_-_17039638 | 1.16 |
ENSDART00000048823
|
c8g
|
complement component 8, gamma polypeptide |
| chr8_+_48613040 | 1.15 |
ENSDART00000121432
|
nppa
|
natriuretic peptide A |
| chr17_-_32863250 | 1.14 |
ENSDART00000167292
|
prox1a
|
prospero homeobox 1a |
| chr8_+_15254564 | 1.14 |
ENSDART00000024433
|
slc5a9
|
solute carrier family 5 (sodium/sugar cotransporter), member 9 |
| chr1_+_47446032 | 1.13 |
ENSDART00000126904
ENSDART00000007262 |
gja8b
|
gap junction protein alpha 8 paralog b |
| chr9_-_48370645 | 1.13 |
ENSDART00000140185
|
col28a2a
|
collagen, type XXVIII, alpha 2a |
| chr9_-_34296406 | 1.13 |
ENSDART00000125451
|
ildr2
|
immunoglobulin-like domain containing receptor 2 |
| chr18_-_21271373 | 1.11 |
ENSDART00000060001
|
pnp6
|
purine nucleoside phosphorylase 6 |
| chr17_+_38573471 | 1.09 |
ENSDART00000040627
|
sptb
|
spectrin, beta, erythrocytic |
| chr17_-_30666037 | 1.09 |
ENSDART00000156509
|
alkal2b
|
ALK and LTK ligand 2b |
| chr10_+_21576909 | 1.09 |
ENSDART00000168604
ENSDART00000166533 |
pcdh1a3
|
protocadherin 1 alpha 3 |
| chr3_-_51912019 | 1.09 |
ENSDART00000149914
|
aatka
|
apoptosis-associated tyrosine kinase a |
| chr21_-_22730832 | 1.08 |
ENSDART00000101797
|
fbxo40.1
|
F-box protein 40, tandem duplicate 1 |
| chr3_-_50046004 | 1.07 |
ENSDART00000109544
|
si:ch1073-100f3.2
|
si:ch1073-100f3.2 |
| chr19_-_10243148 | 1.06 |
ENSDART00000148073
|
shisa7b
|
shisa family member 7 |
| chr7_-_27037990 | 1.06 |
ENSDART00000173561
|
nucb2a
|
nucleobindin 2a |
| chr12_-_31009315 | 1.06 |
ENSDART00000133854
|
tcf7l2
|
transcription factor 7 like 2 |
| chr8_-_25980694 | 1.05 |
ENSDART00000135456
|
si:dkey-72l14.3
|
si:dkey-72l14.3 |
| chr1_+_37391141 | 1.04 |
ENSDART00000083593
ENSDART00000168647 |
sparcl1
|
SPARC-like 1 |
| chr5_+_53482597 | 1.04 |
ENSDART00000180333
|
BX323994.1
|
|
| chr18_+_22994427 | 1.03 |
ENSDART00000173131
ENSDART00000172995 ENSDART00000172923 ENSDART00000141521 ENSDART00000173201 ENSDART00000059961 |
cbfb
|
core-binding factor, beta subunit |
| chr14_-_7128980 | 1.03 |
ENSDART00000171311
|
si:ch73-43g23.1
|
si:ch73-43g23.1 |
| chr21_-_4032650 | 1.02 |
ENSDART00000151648
|
ntng2b
|
netrin g2b |
| chr23_-_24148646 | 1.02 |
ENSDART00000113598
|
arhgef19
|
Rho guanine nucleotide exchange factor (GEF) 19 |
| chr25_-_10503043 | 1.01 |
ENSDART00000155404
|
cox8b
|
cytochrome c oxidase subunit 8b |
| chr7_-_27686021 | 1.01 |
ENSDART00000079112
ENSDART00000100989 |
calca
|
calcitonin/calcitonin-related polypeptide, alpha |
| chr9_-_1949915 | 1.01 |
ENSDART00000190712
|
hoxd3a
|
homeobox D3a |
| chr14_-_26704829 | 1.01 |
ENSDART00000078563
|
neurog1
|
neurogenin 1 |
| chr3_+_15176737 | 1.00 |
ENSDART00000065805
|
tspan10
|
tetraspanin 10 |
| chr7_-_50517023 | 0.99 |
ENSDART00000073910
|
adamtsl5
|
ADAMTS like 5 |
| chr2_-_8017579 | 0.98 |
ENSDART00000040209
|
ephb3a
|
eph receptor B3a |
| chr8_+_29593986 | 0.98 |
ENSDART00000077642
|
atoh1a
|
atonal bHLH transcription factor 1a |
| chr7_+_54605640 | 0.98 |
ENSDART00000168474
|
fgf3
|
fibroblast growth factor 3 |
| chr6_+_21202639 | 0.98 |
ENSDART00000083126
|
cidec
|
cell death-inducing DFFA-like effector c |
| chr18_+_30508729 | 0.98 |
ENSDART00000185140
|
cox4i1
|
cytochrome c oxidase subunit IV isoform 1 |
| chr3_-_32541033 | 0.97 |
ENSDART00000151476
ENSDART00000055324 |
rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr3_-_58644920 | 0.97 |
ENSDART00000155953
|
dhrs7ca
|
dehydrogenase/reductase (SDR family) member 7Ca |
| chr1_+_40801696 | 0.97 |
ENSDART00000147497
|
cpz
|
carboxypeptidase Z |
| chr23_-_4855122 | 0.97 |
ENSDART00000133701
|
slc6a1a
|
solute carrier family 6 (neurotransmitter transporter), member 1a |
| chr9_-_6372535 | 0.97 |
ENSDART00000149189
|
ecrg4a
|
esophageal cancer related gene 4a |
| chr12_+_26471712 | 0.96 |
ENSDART00000162115
|
ndel1a
|
nudE neurodevelopment protein 1-like 1a |
| chr7_+_65939098 | 0.96 |
ENSDART00000193599
|
RASSF10
|
Ras association domain family member 10 |
| chr12_-_31012741 | 0.96 |
ENSDART00000145967
|
tcf7l2
|
transcription factor 7 like 2 |
| chr9_+_21151138 | 0.95 |
ENSDART00000133903
|
hao2
|
hydroxyacid oxidase 2 (long chain) |
| chr1_-_9644630 | 0.95 |
ENSDART00000123725
ENSDART00000161164 |
ugt5b3
|
UDP glucuronosyltransferase 5 family, polypeptide B3 |
| chr14_+_36886950 | 0.95 |
ENSDART00000183719
|
si:ch211-132p1.3
|
si:ch211-132p1.3 |
| chr8_+_8468139 | 0.95 |
ENSDART00000186684
|
comta
|
catechol-O-methyltransferase a |
| chr17_+_42842632 | 0.95 |
ENSDART00000155547
ENSDART00000126087 |
qpct
|
glutaminyl-peptide cyclotransferase |
| chr6_-_24301324 | 0.94 |
ENSDART00000171401
|
BX927081.1
|
|
| chr5_+_52067723 | 0.94 |
ENSDART00000166902
|
setbp1
|
SET binding protein 1 |
| chr16_+_12521872 | 0.94 |
ENSDART00000114411
|
rasip1
|
Ras interacting protein 1 |
| chr21_+_45733871 | 0.94 |
ENSDART00000187285
ENSDART00000193018 |
zgc:77058
|
zgc:77058 |
| chr8_+_8973425 | 0.93 |
ENSDART00000066107
|
bcap31
|
B cell receptor associated protein 31 |
| chr21_+_22630627 | 0.93 |
ENSDART00000193092
|
si:dkeyp-69c1.7
|
si:dkeyp-69c1.7 |
| chr16_+_1254390 | 0.91 |
ENSDART00000092627
|
ADAMTSL4
|
ADAMTS like 4 |
| chr4_+_74131530 | 0.90 |
ENSDART00000174125
|
CABZ01072523.1
|
|
| chr16_+_5579744 | 0.90 |
ENSDART00000147973
|
macf1b
|
microtubule-actin crosslinking factor 1 b |
| chr19_-_24443867 | 0.90 |
ENSDART00000163763
ENSDART00000043133 |
thbs3b
|
thrombospondin 3b |
Network of associatons between targets according to the STRING database.
First level regulatory network of nfia
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 4.0 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) |
| 0.9 | 3.8 | GO:0035630 | bone mineralization involved in bone maturation(GO:0035630) |
| 0.9 | 2.7 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) |
| 0.9 | 3.6 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.7 | 2.2 | GO:0060838 | lymphatic endothelial cell fate commitment(GO:0060838) venous endothelial cell differentiation(GO:0060843) venous endothelial cell fate commitment(GO:0060845) |
| 0.7 | 2.6 | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling(GO:0010882) |
| 0.6 | 1.9 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.6 | 3.1 | GO:0071480 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.5 | 1.6 | GO:0014814 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) |
| 0.5 | 4.3 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.5 | 2.1 | GO:0035790 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) |
| 0.5 | 2.0 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.5 | 1.5 | GO:0033335 | anal fin development(GO:0033335) |
| 0.5 | 1.4 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) symmetric cell division(GO:0098725) |
| 0.5 | 3.3 | GO:0009136 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.5 | 1.4 | GO:0097377 | interneuron axon guidance(GO:0097376) spinal cord interneuron axon guidance(GO:0097377) dorsal spinal cord interneuron axon guidance(GO:0097378) |
| 0.4 | 2.2 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.4 | 1.3 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.4 | 1.3 | GO:2000257 | regulation of protein activation cascade(GO:2000257) |
| 0.4 | 1.6 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.4 | 2.0 | GO:0072575 | hepatocyte proliferation(GO:0072574) epithelial cell proliferation involved in liver morphogenesis(GO:0072575) |
| 0.4 | 1.1 | GO:1905072 | cardiac jelly development(GO:1905072) |
| 0.3 | 1.0 | GO:0021985 | neurohypophysis development(GO:0021985) |
| 0.3 | 1.0 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.3 | 2.1 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.3 | 0.9 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.3 | 0.9 | GO:0021961 | posterior commissure morphogenesis(GO:0021961) |
| 0.3 | 0.9 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.3 | 1.2 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.3 | 1.5 | GO:0014826 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.3 | 0.8 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.3 | 1.3 | GO:0097272 | ammonia homeostasis(GO:0097272) |
| 0.3 | 1.1 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.3 | 1.3 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.3 | 2.8 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.3 | 1.0 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.2 | 0.7 | GO:0071314 | response to cocaine(GO:0042220) cellular response to alkaloid(GO:0071312) cellular response to cocaine(GO:0071314) |
| 0.2 | 2.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.2 | 5.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.2 | 0.7 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.2 | 0.9 | GO:0018199 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) peptidyl-glutamine modification(GO:0018199) |
| 0.2 | 0.9 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.2 | 2.9 | GO:0006603 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.2 | 1.1 | GO:0070378 | ERK5 cascade(GO:0070375) regulation of ERK5 cascade(GO:0070376) positive regulation of ERK5 cascade(GO:0070378) |
| 0.2 | 1.1 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.2 | 1.1 | GO:0015744 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.2 | 1.9 | GO:2001240 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.2 | 0.6 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.2 | 1.3 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.2 | 0.6 | GO:1903673 | mitotic cytokinetic process(GO:1902410) mitotic cleavage furrow formation(GO:1903673) |
| 0.2 | 0.9 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.2 | 0.7 | GO:0014909 | smooth muscle cell migration(GO:0014909) |
| 0.2 | 1.3 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.2 | 1.3 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.2 | 0.7 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.2 | 1.7 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.2 | 0.9 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.2 | 1.0 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.2 | 0.6 | GO:0015859 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.2 | 0.8 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.2 | 0.9 | GO:0019614 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.2 | 1.4 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.2 | 2.0 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.4 | GO:1901232 | regulation of convergent extension involved in axis elongation(GO:1901232) positive regulation of epiboly involved in gastrulation with mouth forming second(GO:1904088) |
| 0.1 | 7.6 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 0.7 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.1 | 0.4 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.1 | 1.9 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 1.5 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.1 | 0.5 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 0.4 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.1 | 0.6 | GO:0034695 | response to prostaglandin E(GO:0034695) cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.1 | 3.5 | GO:0019835 | cytolysis(GO:0019835) |
| 0.1 | 1.7 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.1 | 1.9 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.1 | 1.2 | GO:0032099 | negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.1 | 1.9 | GO:0036065 | fucosylation(GO:0036065) |
| 0.1 | 1.1 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 0.8 | GO:0021570 | rhombomere 4 development(GO:0021570) rhombomere 4 morphogenesis(GO:0021661) |
| 0.1 | 0.2 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.1 | 3.9 | GO:0050654 | chondroitin sulfate proteoglycan metabolic process(GO:0050654) |
| 0.1 | 4.3 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 0.6 | GO:0035176 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.1 | 0.7 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.1 | 0.8 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.1 | 1.7 | GO:0021984 | adenohypophysis development(GO:0021984) |
| 0.1 | 0.4 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.1 | 0.6 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 0.3 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.1 | 1.6 | GO:0033077 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.1 | 0.7 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.1 | 0.8 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.1 | 1.9 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.1 | 0.4 | GO:0010693 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.1 | 0.9 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 | 0.8 | GO:0097090 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.1 | 0.1 | GO:0002090 | regulation of receptor internalization(GO:0002090) |
| 0.1 | 0.5 | GO:0010801 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.1 | 0.8 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.1 | 0.4 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.1 | 0.4 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.1 | 0.7 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.1 | 0.2 | GO:0015874 | norepinephrine transport(GO:0015874) |
| 0.1 | 0.8 | GO:0070570 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.1 | 0.5 | GO:0042403 | thyroid hormone metabolic process(GO:0042403) |
| 0.1 | 0.9 | GO:0090109 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) regulation of focal adhesion assembly(GO:0051893) regulation of cell-substrate junction assembly(GO:0090109) |
| 0.1 | 0.4 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.1 | 0.4 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.1 | 0.4 | GO:0099612 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.1 | 0.9 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.1 | 0.2 | GO:0060823 | canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060823) regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.1 | 0.1 | GO:0032095 | regulation of response to food(GO:0032095) |
| 0.1 | 0.2 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.1 | 0.5 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.1 | 0.7 | GO:0070307 | lens fiber cell development(GO:0070307) lens fiber cell morphogenesis(GO:0070309) |
| 0.1 | 1.3 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 0.4 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 | 0.3 | GO:0071938 | vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.1 | 0.5 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 1.9 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.6 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 1.5 | GO:0007379 | somite specification(GO:0001757) segment specification(GO:0007379) |
| 0.1 | 0.1 | GO:0044878 | abscission(GO:0009838) mitotic cytokinesis checkpoint(GO:0044878) |
| 0.1 | 0.4 | GO:0010573 | vascular endothelial growth factor production(GO:0010573) regulation of vascular endothelial growth factor production(GO:0010574) positive regulation of vascular endothelial growth factor production(GO:0010575) esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.1 | 1.6 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.1 | 1.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 2.3 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.1 | 4.3 | GO:0021510 | spinal cord development(GO:0021510) |
| 0.1 | 0.2 | GO:0071435 | potassium ion export(GO:0071435) potassium ion export across plasma membrane(GO:0097623) |
| 0.1 | 0.5 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 0.8 | GO:1900107 | regulation of nodal signaling pathway(GO:1900107) |
| 0.0 | 1.2 | GO:0003171 | atrioventricular valve development(GO:0003171) |
| 0.0 | 0.6 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 2.0 | GO:0060402 | cytosolic calcium ion transport(GO:0060401) calcium ion transport into cytosol(GO:0060402) |
| 0.0 | 0.5 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.0 | 0.5 | GO:0051350 | negative regulation of adenylate cyclase activity(GO:0007194) negative regulation of cyclic nucleotide metabolic process(GO:0030800) negative regulation of cyclic nucleotide biosynthetic process(GO:0030803) negative regulation of nucleotide biosynthetic process(GO:0030809) negative regulation of cAMP metabolic process(GO:0030815) negative regulation of cAMP biosynthetic process(GO:0030818) negative regulation of cyclase activity(GO:0031280) negative regulation of lyase activity(GO:0051350) negative regulation of purine nucleotide biosynthetic process(GO:1900372) |
| 0.0 | 2.9 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 11.0 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 0.1 | GO:0032814 | natural killer cell activation(GO:0030101) regulation of natural killer cell activation(GO:0032814) positive regulation of natural killer cell activation(GO:0032816) regulation of transforming growth factor beta production(GO:0071634) negative regulation of transforming growth factor beta production(GO:0071635) |
| 0.0 | 4.2 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 0.4 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 0.2 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.0 | 1.3 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.0 | 1.3 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 1.2 | GO:0060840 | artery development(GO:0060840) |
| 0.0 | 0.8 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.2 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 1.2 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 2.9 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.5 | GO:0045807 | positive regulation of endocytosis(GO:0045807) |
| 0.0 | 0.7 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.0 | 0.3 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.3 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 | 0.4 | GO:0042119 | granulocyte activation(GO:0036230) neutrophil activation(GO:0042119) |
| 0.0 | 0.3 | GO:0035141 | medial fin morphogenesis(GO:0035141) |
| 0.0 | 0.8 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.8 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 1.3 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.0 | 0.9 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 1.6 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 1.8 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.6 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) potassium ion homeostasis(GO:0055075) sodium ion export(GO:0071436) |
| 0.0 | 0.3 | GO:0072506 | phosphate ion homeostasis(GO:0055062) trivalent inorganic anion homeostasis(GO:0072506) |
| 0.0 | 2.8 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 0.6 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.6 | GO:0071350 | interleukin-15-mediated signaling pathway(GO:0035723) response to interleukin-15(GO:0070672) cellular response to interleukin-15(GO:0071350) |
| 0.0 | 0.8 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 0.2 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 1.6 | GO:1901214 | regulation of neuron death(GO:1901214) |
| 0.0 | 1.1 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.5 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 2.0 | GO:0016331 | morphogenesis of embryonic epithelium(GO:0016331) |
| 0.0 | 0.1 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.0 | 0.7 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.6 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.1 | GO:0000050 | urea cycle(GO:0000050) urea metabolic process(GO:0019627) |
| 0.0 | 0.6 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 1.2 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.8 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.4 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.0 | 0.5 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.5 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 3.6 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 0.2 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.5 | GO:0019226 | transmission of nerve impulse(GO:0019226) |
| 0.0 | 1.3 | GO:0010770 | positive regulation of cell morphogenesis involved in differentiation(GO:0010770) |
| 0.0 | 3.1 | GO:0072659 | protein localization to plasma membrane(GO:0072659) protein localization to cell periphery(GO:1990778) |
| 0.0 | 0.2 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.0 | 0.5 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 1.0 | GO:0006119 | oxidative phosphorylation(GO:0006119) |
| 0.0 | 0.3 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 0.7 | GO:2000181 | negative regulation of angiogenesis(GO:0016525) negative regulation of blood vessel morphogenesis(GO:2000181) |
| 0.0 | 0.2 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.1 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 1.0 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.1 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 | 0.4 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.5 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 1.2 | GO:0048469 | cell maturation(GO:0048469) |
| 0.0 | 0.3 | GO:0040001 | establishment of mitotic spindle localization(GO:0040001) |
| 0.0 | 0.4 | GO:0030166 | proteoglycan biosynthetic process(GO:0030166) |
| 0.0 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.8 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.6 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.1 | GO:1900744 | regulation of p38MAPK cascade(GO:1900744) |
| 0.0 | 0.1 | GO:0007140 | male meiosis(GO:0007140) |
| 0.0 | 0.4 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.1 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.9 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.6 | 3.0 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.5 | 2.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.4 | 5.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.4 | 2.6 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.3 | 1.9 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.2 | 1.8 | GO:0030428 | cell septum(GO:0030428) |
| 0.2 | 1.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.2 | 3.5 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.2 | 7.6 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 0.5 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.2 | 1.7 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.2 | 1.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.2 | 0.5 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.1 | 1.9 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 2.2 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.1 | 0.5 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 0.7 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 2.4 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.1 | 0.5 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.1 | 14.0 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 2.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 8.4 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 0.7 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 0.7 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.6 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 1.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.2 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 0.9 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.1 | 1.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 0.8 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 0.4 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 2.8 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 1.0 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 0.4 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 2.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 3.1 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 1.5 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 3.9 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.7 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 1.2 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.8 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.0 | 1.3 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.4 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 1.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.2 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 38.5 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.3 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.5 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 14.9 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 0.9 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 2.0 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.8 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.7 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 1.4 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.7 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 2.6 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 0.2 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.3 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.6 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.9 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.0 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.6 | 2.4 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.4 | 5.1 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.4 | 1.1 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.3 | 1.0 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.3 | 0.9 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.3 | 1.9 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.3 | 1.3 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.3 | 3.1 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.3 | 2.5 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.3 | 1.5 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.3 | 2.2 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.3 | 3.3 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.3 | 1.3 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.2 | 0.7 | GO:0016496 | substance P receptor activity(GO:0016496) |
| 0.2 | 4.9 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.2 | 3.6 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.2 | 0.7 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.2 | 0.9 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.2 | 3.0 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.2 | 1.8 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.2 | 1.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.2 | 2.9 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.2 | 1.1 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) |
| 0.2 | 1.1 | GO:0015131 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.2 | 1.7 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.2 | 7.6 | GO:0005518 | collagen binding(GO:0005518) |
| 0.2 | 1.8 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.2 | 0.6 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.2 | 1.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.2 | 0.7 | GO:0048407 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 0.9 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.2 | 0.5 | GO:0005183 | gonadotropin hormone-releasing hormone activity(GO:0005183) gonadotropin-releasing hormone receptor binding(GO:0031530) |
| 0.2 | 3.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 2.7 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 2.1 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.2 | 1.0 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.2 | 0.8 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.2 | 0.6 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.2 | 1.2 | GO:0036122 | BMP binding(GO:0036122) |
| 0.1 | 0.7 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.1 | 0.4 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.1 | 1.0 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 2.4 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 1.5 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.1 | 1.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 0.4 | GO:0008397 | sterol 12-alpha-hydroxylase activity(GO:0008397) |
| 0.1 | 2.0 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 1.5 | GO:0055102 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.1 | 1.9 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.1 | 3.1 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 0.4 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.1 | 3.0 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 2.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.7 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 4.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.9 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.1 | 0.6 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.1 | 0.5 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 0.7 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 1.3 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 11.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 5.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 1.3 | GO:0043236 | laminin binding(GO:0043236) |
| 0.1 | 0.8 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 0.5 | GO:0019202 | amino acid kinase activity(GO:0019202) |
| 0.1 | 0.4 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 0.8 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.1 | 1.3 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.1 | 1.9 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 1.7 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 0.8 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 0.3 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.1 | 0.4 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.1 | 0.3 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.1 | 0.6 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.1 | 1.5 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 1.7 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 5.1 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 0.9 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 0.4 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.1 | 0.7 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 0.6 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) |
| 0.1 | 0.8 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 18.9 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 0.3 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.3 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 1.1 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.2 | GO:0008459 | chondroitin 6-sulfotransferase activity(GO:0008459) |
| 0.0 | 0.6 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.9 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.3 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.0 | 1.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.6 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 1.0 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 1.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.2 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.0 | 0.2 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.3 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 2.0 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 1.9 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.5 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 2.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.8 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.3 | GO:0048018 | receptor activator activity(GO:0030546) receptor agonist activity(GO:0048018) |
| 0.0 | 0.3 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.6 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.0 | 0.6 | GO:1990782 | protein tyrosine kinase binding(GO:1990782) |
| 0.0 | 2.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.9 | GO:0016229 | steroid dehydrogenase activity(GO:0016229) |
| 0.0 | 0.2 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 0.3 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.5 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.6 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.2 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.4 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 1.0 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.7 | GO:0098632 | protein binding involved in cell adhesion(GO:0098631) protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 11.6 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 0.3 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.4 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 3.9 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.7 | GO:0019209 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.0 | 0.1 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 17.3 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 1.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 1.4 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.0 | GO:0080132 | fatty acid alpha-hydroxylase activity(GO:0080132) |
| 0.0 | 2.5 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.1 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.5 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.5 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.5 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.7 | GO:0051427 | hormone receptor binding(GO:0051427) |
| 0.0 | 1.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.2 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.3 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.2 | GO:0098918 | structural constituent of presynaptic active zone(GO:0098882) structural constituent of synapse(GO:0098918) |
| 0.0 | 0.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 18.4 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.2 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.8 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 7.2 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 1.6 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 1.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 0.8 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 2.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 0.7 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 1.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.8 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.3 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 1.7 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.4 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 2.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.5 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 1.8 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 3.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.9 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.3 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 2.6 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.9 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.5 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.6 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.2 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.5 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.1 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 1.8 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.0 | 0.1 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.5 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.2 | 4.1 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.2 | 1.4 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.2 | 4.6 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 1.0 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.1 | 1.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 2.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 0.9 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 4.8 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 0.4 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 0.6 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 0.7 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 1.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 0.4 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 0.7 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 0.3 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.1 | 1.1 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 0.5 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 0.5 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 0.8 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 2.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.6 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 2.9 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.4 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 2.3 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 1.6 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 1.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.4 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.4 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.6 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.3 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |