Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
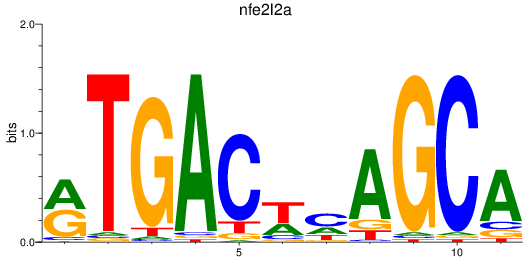
Results for nfe2l2a
Z-value: 1.48
Transcription factors associated with nfe2l2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nfe2l2a
|
ENSDARG00000042824 | nuclear factor, erythroid 2-like 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nfe2l2a | dr11_v1_chr9_+_1654284_1654284 | 0.96 | 4.0e-05 | Click! |
Activity profile of nfe2l2a motif
Sorted Z-values of nfe2l2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_67471375 | 4.52 |
ENSDART00000147009
|
si:dkey-251i10.2
|
si:dkey-251i10.2 |
| chr3_-_26017592 | 3.91 |
ENSDART00000030890
|
hmox1a
|
heme oxygenase 1a |
| chr3_-_26017831 | 3.88 |
ENSDART00000179982
|
hmox1a
|
heme oxygenase 1a |
| chr3_-_55139127 | 3.64 |
ENSDART00000115324
|
hbae1.3
|
hemoglobin, alpha embryonic 1.3 |
| chr9_-_22076368 | 3.22 |
ENSDART00000128486
|
crygm2a
|
crystallin, gamma M2a |
| chr19_-_7450796 | 3.13 |
ENSDART00000104750
|
mllt11
|
MLLT11, transcription factor 7 cofactor |
| chr5_-_30615901 | 3.01 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr19_-_30404096 | 2.64 |
ENSDART00000103475
|
agr2
|
anterior gradient 2 |
| chr21_+_20383837 | 2.23 |
ENSDART00000026430
|
hspb11
|
heat shock protein, alpha-crystallin-related, b11 |
| chr22_-_26595027 | 2.18 |
ENSDART00000184162
|
CABZ01072309.1
|
|
| chr19_+_48176745 | 2.13 |
ENSDART00000164963
|
prdm1b
|
PR domain containing 1b, with ZNF domain |
| chr19_-_30403922 | 2.11 |
ENSDART00000181841
|
agr2
|
anterior gradient 2 |
| chr7_+_38750871 | 2.07 |
ENSDART00000114238
ENSDART00000052325 ENSDART00000137001 |
f2
|
coagulation factor II (thrombin) |
| chr14_+_34780886 | 1.84 |
ENSDART00000130469
ENSDART00000188806 ENSDART00000172924 ENSDART00000173266 ENSDART00000166377 ENSDART00000173371 |
sh3tc2
|
SH3 domain and tetratricopeptide repeats 2 |
| chr14_+_11457500 | 1.81 |
ENSDART00000169202
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr11_+_2198831 | 1.78 |
ENSDART00000160515
|
hoxc6b
|
homeobox C6b |
| chr19_-_6385594 | 1.68 |
ENSDART00000104950
|
atp1a3a
|
ATPase Na+/K+ transporting subunit alpha 3a |
| chr13_+_28903959 | 1.64 |
ENSDART00000166435
|
si:ch73-28h20.1
|
si:ch73-28h20.1 |
| chr13_-_23007813 | 1.62 |
ENSDART00000057638
|
hk1
|
hexokinase 1 |
| chr11_-_24681292 | 1.62 |
ENSDART00000089601
|
olfml3b
|
olfactomedin-like 3b |
| chr3_+_23669267 | 1.55 |
ENSDART00000111227
|
hoxb10a
|
homeobox B10a |
| chr20_-_39103119 | 1.48 |
ENSDART00000143379
|
rcan2
|
regulator of calcineurin 2 |
| chr9_+_41103127 | 1.44 |
ENSDART00000000250
|
slc40a1
|
solute carrier family 40 (iron-regulated transporter), member 1 |
| chr12_+_48216662 | 1.34 |
ENSDART00000187369
|
lrrc20
|
leucine rich repeat containing 20 |
| chr7_-_15413019 | 1.29 |
ENSDART00000172147
|
si:ch211-106j24.1
|
si:ch211-106j24.1 |
| chr13_-_21688176 | 1.26 |
ENSDART00000063825
|
sprn
|
shadow of prion protein |
| chr3_-_21280373 | 1.24 |
ENSDART00000003939
|
syngr1a
|
synaptogyrin 1a |
| chr10_-_7974155 | 1.24 |
ENSDART00000147368
ENSDART00000075524 |
osbp2
|
oxysterol binding protein 2 |
| chr11_-_25418856 | 1.23 |
ENSDART00000013714
|
gata1a
|
GATA binding protein 1a |
| chr11_-_3334248 | 1.18 |
ENSDART00000154314
ENSDART00000121861 |
prph
|
peripherin |
| chr5_-_40297003 | 1.13 |
ENSDART00000097526
|
hspb3
|
heat shock protein, alpha-crystallin-related, b3 |
| chr11_+_26609110 | 1.12 |
ENSDART00000042322
|
map1lc3a
|
microtubule-associated protein 1 light chain 3 alpha |
| chr5_-_64823750 | 1.11 |
ENSDART00000140305
|
lix1
|
limb and CNS expressed 1 |
| chr14_-_17588345 | 1.10 |
ENSDART00000143486
|
selenot2
|
selenoprotein T, 2 |
| chr12_-_1951233 | 1.09 |
ENSDART00000005676
ENSDART00000127937 |
sox9a
|
SRY (sex determining region Y)-box 9a |
| chr14_-_25985698 | 1.09 |
ENSDART00000172909
ENSDART00000123053 |
atox1
|
antioxidant 1 copper chaperone |
| chr7_-_39552314 | 1.06 |
ENSDART00000134174
|
slc22a18
|
solute carrier family 22, member 18 |
| chr5_-_44843738 | 1.00 |
ENSDART00000003926
|
fbp1a
|
fructose-1,6-bisphosphatase 1a |
| chr10_+_29265463 | 0.99 |
ENSDART00000155390
|
crebzf
|
CREB/ATF bZIP transcription factor |
| chr24_-_24146875 | 0.98 |
ENSDART00000173052
|
map7d2b
|
MAP7 domain containing 2b |
| chr1_-_40227166 | 0.97 |
ENSDART00000146680
|
si:ch211-113e8.3
|
si:ch211-113e8.3 |
| chr25_+_34581494 | 0.96 |
ENSDART00000167690
|
trpm1b
|
transient receptor potential cation channel, subfamily M, member 1b |
| chr22_-_31059670 | 0.94 |
ENSDART00000022445
|
cand2
|
cullin-associated and neddylation-dissociated 2 (putative) |
| chr21_+_45819662 | 0.94 |
ENSDART00000193362
ENSDART00000184255 |
pitx1
|
paired-like homeodomain 1 |
| chr7_+_63325819 | 0.94 |
ENSDART00000085612
ENSDART00000161436 |
pcdh7b
|
protocadherin 7b |
| chr7_-_32833153 | 0.94 |
ENSDART00000099871
ENSDART00000099872 |
slc17a6b
|
solute carrier family 17 (vesicular glutamate transporter), member 6b |
| chr1_-_5745420 | 0.93 |
ENSDART00000166779
|
nrp2a
|
neuropilin 2a |
| chr20_-_39271844 | 0.93 |
ENSDART00000192708
|
clu
|
clusterin |
| chr6_-_55958705 | 0.92 |
ENSDART00000155963
|
eya2
|
EYA transcriptional coactivator and phosphatase 2 |
| chr6_-_35446110 | 0.92 |
ENSDART00000058773
|
rgs16
|
regulator of G protein signaling 16 |
| chr4_-_16334362 | 0.89 |
ENSDART00000101461
|
epyc
|
epiphycan |
| chr14_+_30279391 | 0.87 |
ENSDART00000172794
|
fgl1
|
fibrinogen-like 1 |
| chr22_-_13857729 | 0.86 |
ENSDART00000177971
|
s100b
|
S100 calcium binding protein, beta (neural) |
| chr17_+_45106424 | 0.86 |
ENSDART00000157338
ENSDART00000155104 |
GSTZ1
|
zgc:163014 |
| chr13_+_35746440 | 0.85 |
ENSDART00000187859
|
gpr75
|
G protein-coupled receptor 75 |
| chr19_+_8506178 | 0.84 |
ENSDART00000189689
|
s100a10a
|
S100 calcium binding protein A10a |
| chr21_+_5993188 | 0.83 |
ENSDART00000048399
|
slc4a4b
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4b |
| chr8_-_34051548 | 0.83 |
ENSDART00000105204
|
pbx3b
|
pre-B-cell leukemia homeobox 3b |
| chr8_-_34052019 | 0.83 |
ENSDART00000040126
ENSDART00000159208 ENSDART00000048994 ENSDART00000098822 |
pbx3b
|
pre-B-cell leukemia homeobox 3b |
| chr9_-_23253870 | 0.81 |
ENSDART00000143657
ENSDART00000169911 |
acmsd
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr1_+_52398205 | 0.81 |
ENSDART00000143225
|
si:ch211-217k17.9
|
si:ch211-217k17.9 |
| chr7_+_57109214 | 0.80 |
ENSDART00000135068
ENSDART00000098412 |
enosf1
|
enolase superfamily member 1 |
| chr19_-_17658160 | 0.78 |
ENSDART00000151766
ENSDART00000170790 ENSDART00000186678 ENSDART00000188045 ENSDART00000176980 ENSDART00000166313 ENSDART00000188589 |
thrb
|
thyroid hormone receptor beta |
| chr21_-_41025340 | 0.77 |
ENSDART00000148231
|
plac8l1
|
PLAC8-like 1 |
| chr23_-_29376859 | 0.76 |
ENSDART00000146411
|
sst6
|
somatostatin 6 |
| chr24_+_41940299 | 0.75 |
ENSDART00000022349
|
epb41l3a
|
erythrocyte membrane protein band 4.1-like 3a |
| chr7_-_39551913 | 0.75 |
ENSDART00000173498
ENSDART00000143040 |
slc22a18
|
solute carrier family 22, member 18 |
| chr17_-_17948587 | 0.75 |
ENSDART00000090447
|
hhipl1
|
HHIP-like 1 |
| chr14_-_17072736 | 0.74 |
ENSDART00000106333
|
phox2bb
|
paired-like homeobox 2bb |
| chr24_-_16917086 | 0.74 |
ENSDART00000110715
|
cmbl
|
carboxymethylenebutenolidase homolog (Pseudomonas) |
| chr20_-_16906623 | 0.73 |
ENSDART00000012859
ENSDART00000171628 |
psma6b
|
proteasome subunit alpha 6b |
| chr15_+_37559570 | 0.71 |
ENSDART00000085522
|
hspb6
|
heat shock protein, alpha-crystallin-related, b6 |
| chr16_+_13855039 | 0.70 |
ENSDART00000113764
ENSDART00000143983 |
zgc:174888
|
zgc:174888 |
| chr22_-_10354381 | 0.69 |
ENSDART00000092050
|
stab1
|
stabilin 1 |
| chr17_+_27176243 | 0.68 |
ENSDART00000162527
|
si:ch211-160f23.7
|
si:ch211-160f23.7 |
| chr20_-_48604621 | 0.67 |
ENSDART00000161769
ENSDART00000157871 |
mgst3a
|
microsomal glutathione S-transferase 3a |
| chr7_-_15413188 | 0.66 |
ENSDART00000185980
|
si:ch211-106j24.1
|
si:ch211-106j24.1 |
| chr16_-_27640995 | 0.66 |
ENSDART00000019658
|
nacad
|
NAC alpha domain containing |
| chr9_+_11202552 | 0.65 |
ENSDART00000151931
|
asic4a
|
acid-sensing (proton-gated) ion channel family member 4a |
| chr16_-_30880236 | 0.64 |
ENSDART00000035583
|
dennd3b
|
DENN/MADD domain containing 3b |
| chr11_-_29936755 | 0.64 |
ENSDART00000145251
|
pir
|
pirin |
| chr5_-_71460556 | 0.63 |
ENSDART00000108804
|
brinp1
|
bone morphogenetic protein/retinoic acid inducible neural-specific 1 |
| chr21_+_9576176 | 0.62 |
ENSDART00000161289
ENSDART00000159899 ENSDART00000162834 |
mapk10
|
mitogen-activated protein kinase 10 |
| chr24_+_25259154 | 0.61 |
ENSDART00000171125
|
gabrr3b
|
gamma-aminobutyric acid (GABA) A receptor, rho 3b |
| chr23_+_3731375 | 0.60 |
ENSDART00000141782
|
smim29
|
small integral membrane protein 29 |
| chr4_+_25630555 | 0.60 |
ENSDART00000133425
|
acot15
|
acyl-CoA thioesterase 15 |
| chr2_-_6115688 | 0.60 |
ENSDART00000081663
|
prdx1
|
peroxiredoxin 1 |
| chr20_-_48604199 | 0.60 |
ENSDART00000161762
ENSDART00000170894 |
mgst3a
|
microsomal glutathione S-transferase 3a |
| chr20_+_38724575 | 0.60 |
ENSDART00000015095
ENSDART00000152972 |
uts1
|
urotensin 1 |
| chr24_+_18299175 | 0.59 |
ENSDART00000140994
|
tpk1
|
thiamin pyrophosphokinase 1 |
| chr16_-_17162485 | 0.59 |
ENSDART00000123011
|
iffo1b
|
intermediate filament family orphan 1b |
| chr18_-_19405616 | 0.59 |
ENSDART00000191290
ENSDART00000090855 |
megf11
|
multiple EGF-like-domains 11 |
| chr10_-_4375190 | 0.59 |
ENSDART00000016102
|
CABZ01073795.1
|
|
| chr4_+_13696537 | 0.59 |
ENSDART00000109195
ENSDART00000122041 ENSDART00000192554 |
nrcama
|
neuronal cell adhesion molecule a |
| chr2_+_9821757 | 0.59 |
ENSDART00000018408
ENSDART00000141227 ENSDART00000144681 ENSDART00000148227 |
anxa13l
|
annexin A13, like |
| chr24_+_18299506 | 0.58 |
ENSDART00000172056
|
tpk1
|
thiamin pyrophosphokinase 1 |
| chr11_+_10548171 | 0.57 |
ENSDART00000191497
|
b3gnt5a
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5a |
| chr7_+_756942 | 0.57 |
ENSDART00000152224
|
zgc:63470
|
zgc:63470 |
| chr14_-_30960470 | 0.56 |
ENSDART00000129989
|
si:ch211-191o15.6
|
si:ch211-191o15.6 |
| chr23_-_18286822 | 0.55 |
ENSDART00000136672
|
fam19a1a
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A1a |
| chr2_+_25278107 | 0.55 |
ENSDART00000131977
|
ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr7_+_62004048 | 0.54 |
ENSDART00000181818
|
smim20
|
small integral membrane protein 20 |
| chr19_+_31404686 | 0.54 |
ENSDART00000078459
|
pip4p2
|
phosphatidylinositol-4,5-bisphosphate 4-phosphatase 2 |
| chr10_+_26747755 | 0.54 |
ENSDART00000100329
|
f9b
|
coagulation factor IXb |
| chr6_+_43903209 | 0.54 |
ENSDART00000006435
|
gpr27
|
G protein-coupled receptor 27 |
| chr17_-_29902187 | 0.54 |
ENSDART00000009104
|
esrrb
|
estrogen-related receptor beta |
| chr2_-_6115389 | 0.53 |
ENSDART00000134921
|
prdx1
|
peroxiredoxin 1 |
| chr10_+_10788811 | 0.53 |
ENSDART00000101077
ENSDART00000139143 |
ptgdsa
|
prostaglandin D2 synthase a |
| chr22_+_19640309 | 0.52 |
ENSDART00000061725
ENSDART00000140819 |
rgmd
|
RGM domain family, member D |
| chr21_-_39327223 | 0.52 |
ENSDART00000115097
|
aifm5
|
apoptosis-inducing factor, mitochondrion-associated, 5 |
| chr5_+_65946222 | 0.51 |
ENSDART00000190969
ENSDART00000161578 |
mymk
|
myomaker, myoblast fusion factor |
| chr3_+_26064091 | 0.51 |
ENSDART00000143697
|
si:dkeyp-69e1.8
|
si:dkeyp-69e1.8 |
| chr8_-_50259448 | 0.51 |
ENSDART00000146056
|
nkx3-1
|
NK3 homeobox 1 |
| chr14_-_34771371 | 0.50 |
ENSDART00000160598
ENSDART00000150413 ENSDART00000168910 |
ablim3
|
actin binding LIM protein family, member 3 |
| chr7_+_38936132 | 0.49 |
ENSDART00000173945
|
creb3l1
|
cAMP responsive element binding protein 3-like 1 |
| chr19_-_7420867 | 0.49 |
ENSDART00000081741
|
rab25a
|
RAB25, member RAS oncogene family a |
| chr19_+_26828502 | 0.48 |
ENSDART00000183654
ENSDART00000126006 ENSDART00000185341 |
FP085398.1
|
|
| chr11_-_23501467 | 0.47 |
ENSDART00000169066
|
plekha6
|
pleckstrin homology domain containing, family A member 6 |
| chr20_-_1314537 | 0.46 |
ENSDART00000144288
|
pcmt
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr19_-_32600823 | 0.46 |
ENSDART00000134149
ENSDART00000187858 |
zgc:91944
|
zgc:91944 |
| chr18_-_5692292 | 0.46 |
ENSDART00000121503
|
cplx3b
|
complexin 3b |
| chr7_-_52842605 | 0.46 |
ENSDART00000083002
|
map1aa
|
microtubule-associated protein 1Aa |
| chr21_-_5393125 | 0.45 |
ENSDART00000146061
|
psmd5
|
proteasome 26S subunit, non-ATPase 5 |
| chr21_-_11820379 | 0.44 |
ENSDART00000126640
|
RHOBTB3
|
si:dkey-6b12.5 |
| chr15_-_39820491 | 0.44 |
ENSDART00000097134
|
robo1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr22_-_26323893 | 0.44 |
ENSDART00000105099
|
capn1b
|
calpain 1, (mu/I) large subunit b |
| chr5_-_65262260 | 0.43 |
ENSDART00000167886
|
col27a1b
|
collagen, type XXVII, alpha 1b |
| chr20_-_16972351 | 0.41 |
ENSDART00000148312
ENSDART00000186702 |
si:ch73-74h11.1
|
si:ch73-74h11.1 |
| chr3_+_36160979 | 0.41 |
ENSDART00000038525
|
si:ch211-234h8.7
|
si:ch211-234h8.7 |
| chr14_-_30490465 | 0.40 |
ENSDART00000173107
|
micu3b
|
mitochondrial calcium uptake family, member 3b |
| chr8_+_29963024 | 0.40 |
ENSDART00000149372
|
ptch1
|
patched 1 |
| chr5_-_54495678 | 0.40 |
ENSDART00000181970
|
alad
|
aminolevulinate dehydratase |
| chr7_+_17992640 | 0.40 |
ENSDART00000173679
|
ehbp1l1a
|
EH domain binding protein 1-like 1a |
| chr14_-_38873095 | 0.39 |
ENSDART00000047050
ENSDART00000173285 ENSDART00000147521 |
gsr
|
glutathione reductase |
| chr18_+_24922125 | 0.39 |
ENSDART00000180385
|
rgma
|
repulsive guidance molecule family member a |
| chr7_-_30127082 | 0.38 |
ENSDART00000173749
|
alpk3b
|
alpha-kinase 3b |
| chr14_-_32884138 | 0.37 |
ENSDART00000105726
|
slc25a5
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 |
| chr2_-_3038904 | 0.37 |
ENSDART00000186795
|
guk1a
|
guanylate kinase 1a |
| chr1_-_52498146 | 0.36 |
ENSDART00000122217
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr3_+_17846890 | 0.36 |
ENSDART00000193384
|
znf385c
|
zinc finger protein 385C |
| chr20_-_1314355 | 0.36 |
ENSDART00000152436
|
pcmt
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr19_-_41404870 | 0.35 |
ENSDART00000167772
|
sem1
|
SEM1, 26S proteasome complex subunit |
| chr5_-_54424019 | 0.35 |
ENSDART00000169270
ENSDART00000164067 |
coq4
|
coenzyme Q4 homolog (S. cerevisiae) |
| chr2_+_9822319 | 0.35 |
ENSDART00000144078
ENSDART00000144371 |
anxa13l
|
annexin A13, like |
| chr21_-_40782393 | 0.35 |
ENSDART00000075808
|
apbb3
|
amyloid beta (A4) precursor protein-binding, family B, member 3 |
| chr6_-_31364475 | 0.34 |
ENSDART00000145715
ENSDART00000134370 |
ak4
|
adenylate kinase 4 |
| chr12_+_20587179 | 0.34 |
ENSDART00000170127
|
arsg
|
arylsulfatase G |
| chr24_+_25258904 | 0.33 |
ENSDART00000155714
|
gabrr3b
|
gamma-aminobutyric acid (GABA) A receptor, rho 3b |
| chr16_+_11029762 | 0.33 |
ENSDART00000091183
|
erfl3
|
Ets2 repressor factor like 3 |
| chr1_+_41609676 | 0.32 |
ENSDART00000183675
|
mogs
|
mannosyl-oligosaccharide glucosidase |
| chr23_+_19558574 | 0.32 |
ENSDART00000137811
|
atp6ap1lb
|
ATPase H+ transporting accessory protein 1 like b |
| chr12_+_5209822 | 0.32 |
ENSDART00000152610
|
SLC35G1
|
si:ch211-197g18.2 |
| chr11_-_25775447 | 0.31 |
ENSDART00000161301
|
plch2b
|
phospholipase C, eta 2b |
| chr20_-_8443425 | 0.31 |
ENSDART00000083908
|
dab1a
|
Dab, reelin signal transducer, homolog 1a (Drosophila) |
| chr18_-_46763170 | 0.31 |
ENSDART00000171880
|
dner
|
delta/notch-like EGF repeat containing |
| chr9_+_28688574 | 0.31 |
ENSDART00000101319
|
zgc:162396
|
zgc:162396 |
| chr24_+_24923166 | 0.30 |
ENSDART00000065288
|
pcyt1ba
|
phosphate cytidylyltransferase 1, choline, beta a |
| chr6_+_20651398 | 0.29 |
ENSDART00000183663
|
slc19a1
|
solute carrier family 19 (folate transporter), member 1 |
| chr23_+_9560797 | 0.29 |
ENSDART00000180014
|
adrm1
|
adhesion regulating molecule 1 |
| chr25_+_32474031 | 0.29 |
ENSDART00000152124
|
sqor
|
sulfide quinone oxidoreductase |
| chr25_+_10485103 | 0.29 |
ENSDART00000104576
|
psmd13
|
proteasome 26S subunit, non-ATPase 13 |
| chr13_+_24519175 | 0.28 |
ENSDART00000190790
|
sertad2b
|
SERTA domain containing 2b |
| chr7_-_25697285 | 0.28 |
ENSDART00000082620
|
dysf
|
dysferlin, limb girdle muscular dystrophy 2B (autosomal recessive) |
| chr4_+_6572364 | 0.28 |
ENSDART00000122574
|
ppp1r3aa
|
protein phosphatase 1, regulatory subunit 3Aa |
| chr14_-_30490763 | 0.28 |
ENSDART00000193166
ENSDART00000183471 ENSDART00000087859 |
micu3b
|
mitochondrial calcium uptake family, member 3b |
| chr23_+_9560991 | 0.28 |
ENSDART00000081433
ENSDART00000131594 ENSDART00000130069 ENSDART00000138601 |
adrm1
|
adhesion regulating molecule 1 |
| chr1_+_17376922 | 0.26 |
ENSDART00000145068
|
fat1a
|
FAT atypical cadherin 1a |
| chr21_+_25092300 | 0.26 |
ENSDART00000168505
|
dixdc1b
|
DIX domain containing 1b |
| chr1_+_9994811 | 0.25 |
ENSDART00000143719
ENSDART00000110749 |
si:dkeyp-75b4.10
|
si:dkeyp-75b4.10 |
| chr17_-_23673864 | 0.25 |
ENSDART00000104738
ENSDART00000128958 |
ptena
|
phosphatase and tensin homolog A |
| chr13_-_17723417 | 0.24 |
ENSDART00000183834
|
vdac2
|
voltage-dependent anion channel 2 |
| chr7_-_2116512 | 0.23 |
ENSDART00000098148
|
si:cabz01007802.1
|
si:cabz01007802.1 |
| chr9_-_33749556 | 0.23 |
ENSDART00000149383
|
ndp
|
Norrie disease (pseudoglioma) |
| chr8_-_38403018 | 0.23 |
ENSDART00000134100
|
sorbs3
|
sorbin and SH3 domain containing 3 |
| chr22_-_18116635 | 0.21 |
ENSDART00000005724
|
ncanb
|
neurocan b |
| chr10_+_38775408 | 0.21 |
ENSDART00000125045
|
dscama
|
Down syndrome cell adhesion molecule a |
| chr24_+_26039464 | 0.21 |
ENSDART00000131017
|
tnk2a
|
tyrosine kinase, non-receptor, 2a |
| chr11_+_13528437 | 0.20 |
ENSDART00000011362
|
arrdc2
|
arrestin domain containing 2 |
| chr23_+_26142807 | 0.20 |
ENSDART00000158878
|
ptpn22
|
protein tyrosine phosphatase, non-receptor type 22 |
| chr24_-_29868151 | 0.20 |
ENSDART00000184802
|
aglb
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase b |
| chr17_-_12966907 | 0.19 |
ENSDART00000022874
|
psma6a
|
proteasome subunit alpha 6a |
| chr17_-_23674065 | 0.17 |
ENSDART00000104735
ENSDART00000182952 |
ptena
|
phosphatase and tensin homolog A |
| chr2_+_37134281 | 0.17 |
ENSDART00000020135
|
pex19
|
peroxisomal biogenesis factor 19 |
| chr11_+_21096339 | 0.17 |
ENSDART00000124574
|
il19l
|
interleukin 19 like |
| chr5_-_50640171 | 0.17 |
ENSDART00000183353
|
mctp1a
|
multiple C2 domains, transmembrane 1a |
| chr19_+_43119698 | 0.16 |
ENSDART00000167847
ENSDART00000186962 ENSDART00000187305 |
eef1a1l1
|
eukaryotic translation elongation factor 1 alpha 1, like 1 |
| chr14_-_38872536 | 0.16 |
ENSDART00000184315
ENSDART00000127479 |
gsr
|
glutathione reductase |
| chr24_+_35911020 | 0.16 |
ENSDART00000088480
|
abcd4
|
ATP-binding cassette, sub-family D (ALD), member 4 |
| chr24_+_24086249 | 0.15 |
ENSDART00000002953
|
lipib
|
lipase, member Ib |
| chr24_+_24086491 | 0.15 |
ENSDART00000145092
|
lipib
|
lipase, member Ib |
| chr14_-_2004291 | 0.15 |
ENSDART00000114039
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr25_-_16818978 | 0.14 |
ENSDART00000104140
|
dyrk4
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chr22_-_557965 | 0.13 |
ENSDART00000001201
|
bysl
|
bystin-like |
| chr14_+_16093889 | 0.13 |
ENSDART00000159181
|
immt
|
inner membrane protein, mitochondrial (mitofilin) |
| chr22_+_558287 | 0.13 |
ENSDART00000189725
|
med20
|
mediator complex subunit 20 |
| chr15_-_8191841 | 0.13 |
ENSDART00000156663
|
bach1a
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 a |
| chr3_+_43374571 | 0.12 |
ENSDART00000182497
|
zfand2a
|
zinc finger, AN1-type domain 2A |
| chr16_+_51326237 | 0.11 |
ENSDART00000168525
|
CU469384.1
|
|
| chr13_+_7241170 | 0.11 |
ENSDART00000109434
|
aifm2
|
apoptosis-inducing factor, mitochondrion-associated, 2 |
| chr9_+_19304697 | 0.11 |
ENSDART00000040449
|
htr1fa
|
5-hydroxytryptamine (serotonin) receptor 1Fa |
Network of associatons between targets according to the STRING database.
First level regulatory network of nfe2l2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 7.8 | GO:0007585 | respiratory gaseous exchange(GO:0007585) regulation of respiratory gaseous exchange(GO:0043576) |
| 0.6 | 3.1 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) regulation of release of cytochrome c from mitochondria(GO:0090199) positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.5 | 4.8 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.5 | 1.9 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.4 | 1.2 | GO:0009229 | thiamine diphosphate biosynthetic process(GO:0009229) |
| 0.4 | 1.4 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.3 | 2.1 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.3 | 1.0 | GO:0005986 | sucrose metabolic process(GO:0005985) sucrose biosynthetic process(GO:0005986) disaccharide biosynthetic process(GO:0046351) |
| 0.3 | 0.9 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.2 | 0.7 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.2 | 0.8 | GO:0046551 | retinal cone cell fate determination(GO:0042671) eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate determination(GO:0043703) retinal cone cell fate commitment(GO:0046551) photoreceptor cell fate commitment(GO:0046552) camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.2 | 4.8 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.2 | 0.7 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.1 | 1.1 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.1 | 0.8 | GO:0046874 | quinolinate biosynthetic process(GO:0019805) quinolinate metabolic process(GO:0046874) |
| 0.1 | 0.5 | GO:0014743 | regulation of muscle hypertrophy(GO:0014743) |
| 0.1 | 0.9 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.1 | 1.7 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 1.6 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.1 | 0.3 | GO:0014014 | negative regulation of gliogenesis(GO:0014014) |
| 0.1 | 0.9 | GO:2001239 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.1 | 0.3 | GO:0098838 | folic acid transport(GO:0015884) drug transport(GO:0015893) methotrexate transport(GO:0051958) reduced folate transmembrane transport(GO:0098838) |
| 0.1 | 2.2 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.1 | 0.3 | GO:0070221 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.1 | 0.4 | GO:0051503 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.1 | 1.7 | GO:0071436 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.1 | 0.4 | GO:0031174 | lifelong otolith mineralization(GO:0031174) |
| 0.1 | 0.4 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 1.1 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) |
| 0.1 | 0.3 | GO:0042117 | plasma membrane repair(GO:0001778) monocyte activation(GO:0042117) |
| 0.1 | 2.2 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.1 | 0.6 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 0.4 | GO:0003262 | endocardial progenitor cell migration to the midline involved in heart field formation(GO:0003262) |
| 0.1 | 0.9 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 1.0 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.1 | 0.8 | GO:0051963 | regulation of synapse assembly(GO:0051963) |
| 0.0 | 0.3 | GO:0009180 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.0 | 0.6 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.4 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.0 | 0.2 | GO:0045046 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.3 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.2 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.0 | 0.9 | GO:0030947 | regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030947) |
| 0.0 | 0.8 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.5 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.2 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 | 1.2 | GO:0045104 | intermediate filament-based process(GO:0045103) intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.4 | GO:0046501 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 1.9 | GO:0008217 | regulation of blood pressure(GO:0008217) |
| 0.0 | 0.5 | GO:0042593 | carbohydrate homeostasis(GO:0033500) glucose homeostasis(GO:0042593) |
| 0.0 | 0.5 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.9 | GO:0050868 | negative regulation of T cell activation(GO:0050868) negative regulation of leukocyte cell-cell adhesion(GO:1903038) |
| 0.0 | 0.4 | GO:0006744 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.5 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 2.2 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.5 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 1.5 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.3 | GO:0010962 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.0 | 1.0 | GO:0060401 | cytosolic calcium ion transport(GO:0060401) calcium ion transport into cytosol(GO:0060402) |
| 0.0 | 0.9 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.2 | GO:0044247 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.3 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.6 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.5 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.9 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.2 | GO:0045454 | cell redox homeostasis(GO:0045454) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.6 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 1.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 0.7 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.1 | 0.9 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 1.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 0.9 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 0.7 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 0.3 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.0 | 0.5 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 1.2 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.4 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.9 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 1.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.5 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 1.8 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.3 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 1.7 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.4 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.4 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.2 | GO:0046930 | pore complex(GO:0046930) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 7.8 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.4 | 1.2 | GO:0030975 | thiamine diphosphokinase activity(GO:0004788) thiamine binding(GO:0030975) |
| 0.3 | 1.0 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.3 | 3.6 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.3 | 1.6 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.2 | 1.5 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.2 | 1.1 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.2 | 0.9 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.2 | 0.8 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.2 | 0.6 | GO:0008457 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.2 | 1.7 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.2 | 1.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.2 | 1.4 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.1 | 0.8 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 1.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.1 | 0.5 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.1 | 0.4 | GO:0051800 | inositol-1,3,4,5-tetrakisphosphate 3-phosphatase activity(GO:0051717) phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.1 | 1.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 0.4 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.1 | 0.3 | GO:0015350 | reduced folate carrier activity(GO:0008518) methotrexate transporter activity(GO:0015350) |
| 0.1 | 0.3 | GO:0070224 | sulfide:quinone oxidoreductase activity(GO:0070224) |
| 0.1 | 0.6 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.4 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.1 | 0.4 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.9 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 0.4 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 0.9 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.1 | 0.5 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.1 | 0.6 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 1.9 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.3 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.8 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 1.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.7 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.2 | GO:0004133 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.0 | 0.2 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.9 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.5 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.3 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.0 | 3.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.4 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.4 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.8 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.3 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.9 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.5 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.3 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.9 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 1.1 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 2.5 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.3 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.3 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.0 | 0.3 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.2 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 1.1 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 1.2 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.6 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.5 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.7 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.6 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.1 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 1.5 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.8 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.6 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.1 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.4 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.4 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.8 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.3 | 2.1 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 0.9 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 1.4 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.1 | 0.6 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 0.4 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.1 | 0.8 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 0.5 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 1.6 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.8 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 1.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 1.5 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.9 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.3 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |