Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
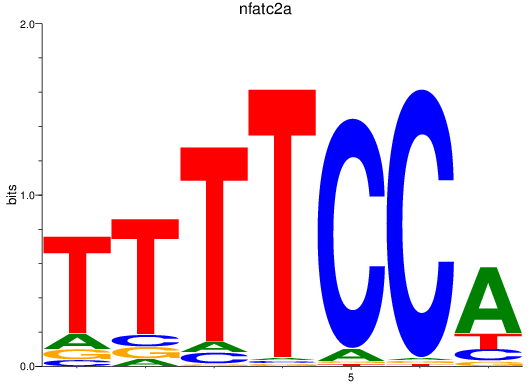
Results for nfatc2a
Z-value: 1.60
Transcription factors associated with nfatc2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nfatc2a
|
ENSDARG00000100927 | nuclear factor of activated T cells 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nfatc2a | dr11_v1_chr23_+_39089574_39089574 | -0.63 | 7.0e-02 | Click! |
Activity profile of nfatc2a motif
Sorted Z-values of nfatc2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_16595177 | 2.92 |
ENSDART00000133962
|
si:dkey-239j18.2
|
si:dkey-239j18.2 |
| chr24_-_28893251 | 2.84 |
ENSDART00000042065
ENSDART00000003503 |
col11a1a
|
collagen, type XI, alpha 1a |
| chr12_-_16595406 | 2.51 |
ENSDART00000166798
|
si:dkey-239j18.2
|
si:dkey-239j18.2 |
| chr16_-_45910050 | 2.38 |
ENSDART00000133213
|
afp4
|
antifreeze protein type IV |
| chr12_-_16941319 | 2.22 |
ENSDART00000109968
|
zgc:174855
|
zgc:174855 |
| chr12_-_16923162 | 2.22 |
ENSDART00000106072
|
si:dkey-26g8.5
|
si:dkey-26g8.5 |
| chr12_-_16558106 | 2.12 |
ENSDART00000109033
|
si:dkey-269i1.4
|
si:dkey-269i1.4 |
| chr23_+_36130883 | 1.97 |
ENSDART00000103132
|
hoxc4a
|
homeobox C4a |
| chr12_-_16898140 | 1.97 |
ENSDART00000152656
|
MGC174155
|
Cathepsin L1-like |
| chr12_+_16949391 | 1.94 |
ENSDART00000152635
|
zgc:174153
|
zgc:174153 |
| chr12_-_16694092 | 1.91 |
ENSDART00000047916
|
ctslb
|
cathepsin Lb |
| chr12_-_16877136 | 1.89 |
ENSDART00000152593
|
si:dkey-269i1.4
|
si:dkey-269i1.4 |
| chr4_-_15420452 | 1.88 |
ENSDART00000016230
|
plxna4
|
plexin A4 |
| chr4_+_7677318 | 1.87 |
ENSDART00000149218
|
elk3
|
ELK3, ETS-domain protein |
| chr3_-_61181018 | 1.84 |
ENSDART00000187970
|
pvalb4
|
parvalbumin 4 |
| chr19_-_41069573 | 1.84 |
ENSDART00000111982
ENSDART00000193142 |
sgce
|
sarcoglycan, epsilon |
| chr3_-_55139127 | 1.84 |
ENSDART00000115324
|
hbae1.3
|
hemoglobin, alpha embryonic 1.3 |
| chr12_-_16720196 | 1.83 |
ENSDART00000187639
|
si:dkey-26g8.4
|
si:dkey-26g8.4 |
| chr16_+_23397785 | 1.82 |
ENSDART00000148961
|
s100a10b
|
S100 calcium binding protein A10b |
| chr6_-_39313027 | 1.81 |
ENSDART00000012644
|
krt4
|
keratin 4 |
| chr14_-_3381303 | 1.76 |
ENSDART00000171601
|
im:7150988
|
im:7150988 |
| chr15_-_106689 | 1.70 |
ENSDART00000185007
|
apoa1b
|
apolipoprotein A-Ib |
| chr2_-_35566938 | 1.68 |
ENSDART00000029006
ENSDART00000077178 ENSDART00000125298 |
tnn
|
tenascin N |
| chr16_+_23984179 | 1.67 |
ENSDART00000175879
|
apoc2
|
apolipoprotein C-II |
| chr12_-_16636627 | 1.60 |
ENSDART00000128811
|
si:dkey-239j18.3
|
si:dkey-239j18.3 |
| chr16_-_14397003 | 1.59 |
ENSDART00000170957
|
crabp2a
|
cellular retinoic acid binding protein 2, a |
| chr12_-_645972 | 1.59 |
ENSDART00000048632
|
sult2st1
|
sulfotransferase family 2, cytosolic sulfotransferase 1 |
| chr12_-_3133483 | 1.54 |
ENSDART00000015092
|
col1a1b
|
collagen, type I, alpha 1b |
| chr22_-_26353916 | 1.51 |
ENSDART00000077958
|
capn2b
|
calpain 2, (m/II) large subunit b |
| chr24_-_21921262 | 1.48 |
ENSDART00000186061
ENSDART00000187846 |
tagln3b
|
transgelin 3b |
| chr15_+_32710300 | 1.48 |
ENSDART00000167857
|
postnb
|
periostin, osteoblast specific factor b |
| chr12_-_16619449 | 1.47 |
ENSDART00000182074
|
ctslb
|
cathepsin Lb |
| chr23_+_36101185 | 1.47 |
ENSDART00000103139
|
hoxc8a
|
homeobox C8a |
| chr12_-_16619757 | 1.46 |
ENSDART00000145570
|
ctslb
|
cathepsin Lb |
| chr1_+_44911405 | 1.44 |
ENSDART00000182465
|
wu:fc21g02
|
wu:fc21g02 |
| chr23_-_20345473 | 1.38 |
ENSDART00000140935
|
si:rp71-17i16.6
|
si:rp71-17i16.6 |
| chr6_-_49526510 | 1.37 |
ENSDART00000128025
|
rps26l
|
ribosomal protein S26, like |
| chr21_+_7582036 | 1.37 |
ENSDART00000135485
ENSDART00000027268 |
otpa
|
orthopedia homeobox a |
| chr2_-_5728843 | 1.37 |
ENSDART00000014020
|
sst2
|
somatostatin 2 |
| chr6_+_15250672 | 1.35 |
ENSDART00000155951
|
si:ch73-23l24.1
|
si:ch73-23l24.1 |
| chr12_-_16720432 | 1.34 |
ENSDART00000152261
ENSDART00000152154 |
si:dkey-26g8.4
|
si:dkey-26g8.4 |
| chr5_-_72125551 | 1.34 |
ENSDART00000149412
|
smyd1a
|
SET and MYND domain containing 1a |
| chr16_-_16182319 | 1.33 |
ENSDART00000103815
|
stmn2a
|
stathmin 2a |
| chr3_+_24197934 | 1.31 |
ENSDART00000055609
|
atf4b
|
activating transcription factor 4b |
| chr20_-_10120442 | 1.29 |
ENSDART00000144970
|
meis2b
|
Meis homeobox 2b |
| chr14_+_6159356 | 1.25 |
ENSDART00000157730
|
bscl2l
|
Bernardinelli-Seip congenital lipodystrophy 2, like |
| chr15_+_33989181 | 1.22 |
ENSDART00000169487
|
vwde
|
von Willebrand factor D and EGF domains |
| chr2_+_22042745 | 1.22 |
ENSDART00000132039
|
tox
|
thymocyte selection-associated high mobility group box |
| chr5_-_65021156 | 1.22 |
ENSDART00000166183
|
anxa1c
|
annexin A1c |
| chr5_+_23242370 | 1.22 |
ENSDART00000051532
|
agtr2
|
angiotensin II receptor, type 2 |
| chr6_-_15653494 | 1.21 |
ENSDART00000038133
|
trim63a
|
tripartite motif containing 63a |
| chr17_+_52822831 | 1.19 |
ENSDART00000193368
|
meis2a
|
Meis homeobox 2a |
| chr5_+_37091626 | 1.19 |
ENSDART00000161054
|
tagln2
|
transgelin 2 |
| chr13_-_30645965 | 1.18 |
ENSDART00000109307
|
zcchc24
|
zinc finger, CCHC domain containing 24 |
| chr1_+_7540978 | 1.18 |
ENSDART00000147770
|
mylz3
|
myosin, light polypeptide 3, skeletal muscle |
| chr25_-_15045338 | 1.17 |
ENSDART00000161165
ENSDART00000165774 ENSDART00000172538 |
pax6a
|
paired box 6a |
| chr5_-_13835461 | 1.17 |
ENSDART00000148297
ENSDART00000114841 |
add2
|
adducin 2 (beta) |
| chr25_+_36152215 | 1.17 |
ENSDART00000036147
|
irx5b
|
iroquois homeobox 5b |
| chr17_+_52822422 | 1.15 |
ENSDART00000158273
ENSDART00000161414 |
meis2a
|
Meis homeobox 2a |
| chr13_-_5568928 | 1.14 |
ENSDART00000192679
|
meis1b
|
Meis homeobox 1 b |
| chr3_+_36275633 | 1.13 |
ENSDART00000185027
ENSDART00000149532 ENSDART00000102883 ENSDART00000148444 |
zgc:86896
|
zgc:86896 |
| chr8_+_17078692 | 1.12 |
ENSDART00000023206
|
plk2b
|
polo-like kinase 2b (Drosophila) |
| chr22_-_4644484 | 1.11 |
ENSDART00000167748
|
fbn2b
|
fibrillin 2b |
| chr7_-_35314347 | 1.10 |
ENSDART00000005053
|
slc12a4
|
solute carrier family 12 (potassium/chloride transporter), member 4 |
| chr7_+_15871156 | 1.10 |
ENSDART00000145946
|
pax6b
|
paired box 6b |
| chr9_-_22831836 | 1.10 |
ENSDART00000142585
|
neb
|
nebulin |
| chr3_-_20091964 | 1.09 |
ENSDART00000029386
ENSDART00000020253 ENSDART00000124326 |
slc4a1a
|
solute carrier family 4 (anion exchanger), member 1a (Diego blood group) |
| chr14_-_39074539 | 1.09 |
ENSDART00000030509
|
glra4a
|
glycine receptor, alpha 4a |
| chr1_-_56223913 | 1.08 |
ENSDART00000019573
|
zgc:65894
|
zgc:65894 |
| chr2_-_716426 | 1.07 |
ENSDART00000028159
|
foxf2a
|
forkhead box F2a |
| chr6_+_43400059 | 1.05 |
ENSDART00000143374
|
mitfa
|
microphthalmia-associated transcription factor a |
| chr2_+_27394798 | 1.04 |
ENSDART00000115071
|
selenop2
|
selenoprotein P2 |
| chr3_+_30921246 | 1.03 |
ENSDART00000076850
|
cldni
|
claudin i |
| chr12_+_18234557 | 1.02 |
ENSDART00000130741
|
fam20cb
|
family with sequence similarity 20, member Cb |
| chr13_+_13681681 | 1.02 |
ENSDART00000057825
|
cfd
|
complement factor D (adipsin) |
| chr17_+_23300827 | 1.01 |
ENSDART00000058745
|
zgc:165461
|
zgc:165461 |
| chr7_+_39401388 | 1.01 |
ENSDART00000144750
|
tnni2b.1
|
troponin I type 2b (skeletal, fast), tandem duplicate 1 |
| chr1_+_44442140 | 1.01 |
ENSDART00000190592
|
crybb1l2
|
crystallin, beta B1, like 2 |
| chr1_+_19303241 | 1.00 |
ENSDART00000129970
|
si:dkeyp-118a3.2
|
si:dkeyp-118a3.2 |
| chr15_+_7176182 | 1.00 |
ENSDART00000101578
|
her8.2
|
hairy-related 8.2 |
| chr2_-_31767827 | 1.00 |
ENSDART00000114928
|
and2
|
actinodin2 |
| chr2_+_42191592 | 1.00 |
ENSDART00000144716
|
cavin4a
|
caveolae associated protein 4a |
| chr6_-_13114821 | 1.00 |
ENSDART00000164579
ENSDART00000165375 |
zgc:194469
|
zgc:194469 |
| chr8_+_46217861 | 1.00 |
ENSDART00000038790
|
angptl7
|
angiopoietin-like 7 |
| chr6_+_2097690 | 0.99 |
ENSDART00000193770
|
tgm2b
|
transglutaminase 2b |
| chr3_-_19899914 | 0.98 |
ENSDART00000134969
|
rnd2
|
Rho family GTPase 2 |
| chr16_+_23398369 | 0.98 |
ENSDART00000037694
|
s100a10b
|
S100 calcium binding protein A10b |
| chr22_+_16022211 | 0.98 |
ENSDART00000062618
|
serpinc1
|
serpin peptidase inhibitor, clade C (antithrombin), member 1 |
| chr24_-_26282672 | 0.98 |
ENSDART00000176663
|
and1
|
actinodin1 |
| chr15_+_29085955 | 0.98 |
ENSDART00000156799
|
si:ch211-137a8.4
|
si:ch211-137a8.4 |
| chr21_+_19445942 | 0.97 |
ENSDART00000030887
|
slc45a2
|
solute carrier family 45, member 2 |
| chr13_-_15994419 | 0.97 |
ENSDART00000079724
ENSDART00000042377 ENSDART00000046079 ENSDART00000050481 ENSDART00000016430 |
ikzf1
|
IKAROS family zinc finger 1 (Ikaros) |
| chr19_+_19772765 | 0.97 |
ENSDART00000182028
ENSDART00000161019 |
hoxa3a
|
homeobox A3a |
| chr12_-_26064480 | 0.96 |
ENSDART00000158215
ENSDART00000171206 ENSDART00000171212 ENSDART00000182956 ENSDART00000186779 |
ldb3b
|
LIM domain binding 3b |
| chr19_-_31522625 | 0.96 |
ENSDART00000158438
ENSDART00000035049 |
necab1
|
N-terminal EF-hand calcium binding protein 1 |
| chr15_+_33991928 | 0.96 |
ENSDART00000170177
|
vwde
|
von Willebrand factor D and EGF domains |
| chr17_+_22102791 | 0.96 |
ENSDART00000047772
|
mal
|
mal, T cell differentiation protein |
| chr21_-_3422635 | 0.95 |
ENSDART00000150975
|
smad7
|
SMAD family member 7 |
| chr2_-_33993533 | 0.95 |
ENSDART00000140910
ENSDART00000077304 |
ptch2
|
patched 2 |
| chr4_+_14360372 | 0.94 |
ENSDART00000007103
|
nuak1a
|
NUAK family, SNF1-like kinase, 1a |
| chr9_+_30090656 | 0.94 |
ENSDART00000102981
|
col8a1a
|
collagen, type VIII, alpha 1a |
| chr11_-_22599584 | 0.93 |
ENSDART00000014062
|
myog
|
myogenin |
| chr14_+_6159162 | 0.93 |
ENSDART00000128638
|
bscl2l
|
Bernardinelli-Seip congenital lipodystrophy 2, like |
| chr16_-_22713152 | 0.93 |
ENSDART00000140953
ENSDART00000143836 |
si:ch211-105c13.3
|
si:ch211-105c13.3 |
| chr2_-_20923864 | 0.92 |
ENSDART00000006870
|
ptgs2a
|
prostaglandin-endoperoxide synthase 2a |
| chr7_+_22657566 | 0.92 |
ENSDART00000141048
|
ponzr5
|
plac8 onzin related protein 5 |
| chr22_-_15593824 | 0.92 |
ENSDART00000123125
|
tpm4a
|
tropomyosin 4a |
| chr3_+_23092762 | 0.91 |
ENSDART00000142884
ENSDART00000024136 |
gngt2a
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2a |
| chr23_+_32499916 | 0.91 |
ENSDART00000134811
|
si:dkey-261h17.1
|
si:dkey-261h17.1 |
| chr4_-_16354292 | 0.91 |
ENSDART00000139919
|
lum
|
lumican |
| chr11_-_3552067 | 0.91 |
ENSDART00000163656
|
CAMK2N1
|
si:dkey-33m11.6 |
| chr24_-_33756003 | 0.90 |
ENSDART00000079283
|
tmeff1b
|
transmembrane protein with EGF-like and two follistatin-like domains 1b |
| chr12_+_7445595 | 0.90 |
ENSDART00000103536
ENSDART00000152524 |
phyhiplb
|
phytanoyl-CoA 2-hydroxylase interacting protein-like b |
| chr2_+_27394979 | 0.90 |
ENSDART00000170495
|
selenop2
|
selenoprotein P2 |
| chr10_+_36650222 | 0.90 |
ENSDART00000126963
|
ucp3
|
uncoupling protein 3 |
| chr10_-_28761454 | 0.89 |
ENSDART00000129400
|
alcama
|
activated leukocyte cell adhesion molecule a |
| chr7_+_38750871 | 0.89 |
ENSDART00000114238
ENSDART00000052325 ENSDART00000137001 |
f2
|
coagulation factor II (thrombin) |
| chr18_-_48530221 | 0.89 |
ENSDART00000188134
ENSDART00000142107 |
kcnj1a.2
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 2 |
| chr13_-_5569562 | 0.89 |
ENSDART00000102576
|
meis1b
|
Meis homeobox 1 b |
| chr11_-_26666501 | 0.89 |
ENSDART00000188067
ENSDART00000111539 |
efcc1
|
EF-hand and coiled-coil domain containing 1 |
| chr12_+_42436920 | 0.88 |
ENSDART00000177303
|
ebf3a
|
early B cell factor 3a |
| chr16_-_45225520 | 0.88 |
ENSDART00000158855
|
fxyd1
|
FXYD domain containing ion transport regulator 1 (phospholemman) |
| chr5_-_28679135 | 0.87 |
ENSDART00000193585
|
tnc
|
tenascin C |
| chr6_-_49063085 | 0.87 |
ENSDART00000156124
|
si:ch211-105j21.9
|
si:ch211-105j21.9 |
| chr5_+_65991152 | 0.87 |
ENSDART00000097756
|
lcn15
|
lipocalin 15 |
| chr6_+_3827751 | 0.86 |
ENSDART00000003008
ENSDART00000122348 |
gad1b
|
glutamate decarboxylase 1b |
| chr18_+_19648275 | 0.86 |
ENSDART00000100569
|
smad6b
|
SMAD family member 6b |
| chr6_-_48094342 | 0.86 |
ENSDART00000137458
|
slc2a1b
|
solute carrier family 2 (facilitated glucose transporter), member 1b |
| chr13_-_39947335 | 0.85 |
ENSDART00000056996
|
sfrp5
|
secreted frizzled-related protein 5 |
| chr18_+_16744307 | 0.85 |
ENSDART00000179872
ENSDART00000133490 |
lyve1b
|
lymphatic vessel endothelial hyaluronic receptor 1b |
| chr20_+_26538137 | 0.85 |
ENSDART00000045397
|
stx11b.1
|
syntaxin 11b, tandem duplicate 1 |
| chr19_-_38611814 | 0.85 |
ENSDART00000151958
|
col16a1
|
collagen, type XVI, alpha 1 |
| chr25_-_22639133 | 0.85 |
ENSDART00000073583
|
islr2
|
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chr25_-_28384954 | 0.85 |
ENSDART00000073500
|
ptprz1a
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1a |
| chr7_+_60551133 | 0.85 |
ENSDART00000148038
|
lrfn4b
|
leucine rich repeat and fibronectin type III domain containing 4b |
| chr7_-_35708450 | 0.84 |
ENSDART00000193886
|
irx5a
|
iroquois homeobox 5a |
| chr7_-_24699985 | 0.84 |
ENSDART00000052802
|
calb2b
|
calbindin 2b |
| chr5_+_29160132 | 0.84 |
ENSDART00000088827
|
dpp7
|
dipeptidyl-peptidase 7 |
| chr4_+_22680442 | 0.84 |
ENSDART00000036531
|
gnai1
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
| chr22_+_31039091 | 0.84 |
ENSDART00000060005
|
rpl32
|
ribosomal protein L32 |
| chr16_-_35975254 | 0.84 |
ENSDART00000167537
|
eva1ba
|
eva-1 homolog Ba (C. elegans) |
| chr8_+_13106760 | 0.83 |
ENSDART00000029308
|
itgb4
|
integrin, beta 4 |
| chr15_+_32710094 | 0.83 |
ENSDART00000159592
|
postnb
|
periostin, osteoblast specific factor b |
| chr11_+_6116503 | 0.83 |
ENSDART00000176170
|
nr2f6b
|
nuclear receptor subfamily 2, group F, member 6b |
| chr10_+_35265083 | 0.83 |
ENSDART00000048831
|
tmem120a
|
transmembrane protein 120A |
| chr18_-_17415580 | 0.82 |
ENSDART00000150077
ENSDART00000100190 |
ces2
|
carboxylesterase 2 (intestine, liver) |
| chr7_-_27037990 | 0.82 |
ENSDART00000173561
|
nucb2a
|
nucleobindin 2a |
| chr25_-_27541621 | 0.82 |
ENSDART00000130678
|
spam1
|
sperm adhesion molecule 1 |
| chr5_+_33339762 | 0.82 |
ENSDART00000026085
|
ptges
|
prostaglandin E synthase |
| chr1_-_12278522 | 0.82 |
ENSDART00000142122
ENSDART00000003825 |
cplx2l
|
complexin 2, like |
| chr25_+_14507567 | 0.82 |
ENSDART00000015681
|
dbx1b
|
developing brain homeobox 1b |
| chr15_+_1372343 | 0.81 |
ENSDART00000152285
|
schip1
|
schwannomin interacting protein 1 |
| chr21_+_5636008 | 0.81 |
ENSDART00000158385
|
shroom3
|
shroom family member 3 |
| chr22_-_23253481 | 0.81 |
ENSDART00000054807
|
lhx9
|
LIM homeobox 9 |
| chr11_-_32723851 | 0.81 |
ENSDART00000155592
|
pcdh17
|
protocadherin 17 |
| chr19_+_12444943 | 0.80 |
ENSDART00000135706
|
ldlrad4a
|
low density lipoprotein receptor class A domain containing 4a |
| chr9_-_1959917 | 0.80 |
ENSDART00000082359
|
hoxd3a
|
homeobox D3a |
| chr20_-_43771871 | 0.80 |
ENSDART00000153304
|
matn3a
|
matrilin 3a |
| chr16_-_24044664 | 0.80 |
ENSDART00000136982
|
bcam
|
basal cell adhesion molecule (Lutheran blood group) |
| chr7_-_7398350 | 0.79 |
ENSDART00000012637
|
zgc:101810
|
zgc:101810 |
| chr9_-_12424791 | 0.79 |
ENSDART00000135447
ENSDART00000088199 |
zgc:162707
|
zgc:162707 |
| chr12_+_17106117 | 0.79 |
ENSDART00000149990
|
acta2
|
actin, alpha 2, smooth muscle, aorta |
| chr6_-_13114406 | 0.78 |
ENSDART00000188015
|
zgc:194469
|
zgc:194469 |
| chr18_+_17663898 | 0.78 |
ENSDART00000021213
|
cpne2
|
copine II |
| chr25_-_37489917 | 0.78 |
ENSDART00000160688
|
psma1
|
proteasome subunit alpha 1 |
| chr23_+_35672542 | 0.77 |
ENSDART00000046268
|
pmelb
|
premelanosome protein b |
| chr19_-_24267410 | 0.77 |
ENSDART00000104083
|
s100v2
|
S100 calcium binding protein V2 |
| chr17_-_36929332 | 0.77 |
ENSDART00000183454
|
dpysl5a
|
dihydropyrimidinase-like 5a |
| chr4_+_47257854 | 0.77 |
ENSDART00000173868
|
crestin
|
crestin |
| chr14_+_8174828 | 0.77 |
ENSDART00000167228
|
psd2
|
pleckstrin and Sec7 domain containing 2 |
| chr9_-_23118350 | 0.76 |
ENSDART00000020884
|
lypd6
|
LY6/PLAUR domain containing 6 |
| chr11_+_25481046 | 0.76 |
ENSDART00000065940
|
opn1lw2
|
opsin 1 (cone pigments), long-wave-sensitive, 2 |
| chr1_-_40911332 | 0.76 |
ENSDART00000027463
|
hmx4
|
H6 family homeobox 4 |
| chr18_-_39583601 | 0.76 |
ENSDART00000125116
|
tnfaip8l3
|
tumor necrosis factor, alpha-induced protein 8-like 3 |
| chr1_+_45351890 | 0.76 |
ENSDART00000145486
|
si:ch211-243a20.3
|
si:ch211-243a20.3 |
| chr22_+_3914318 | 0.76 |
ENSDART00000188774
ENSDART00000082034 |
FO904903.1
|
Danio rerio major histocompatibility complex class I ULA (mhc1ula), mRNA. |
| chr14_+_49251331 | 0.76 |
ENSDART00000148882
|
anxa6
|
annexin A6 |
| chr23_+_36115541 | 0.76 |
ENSDART00000130090
|
hoxc6a
|
homeobox C6a |
| chr10_+_21660447 | 0.76 |
ENSDART00000164519
|
pcdh1g3
|
protocadherin 1 gamma 3 |
| chr25_-_27541288 | 0.75 |
ENSDART00000187245
|
spam1
|
sperm adhesion molecule 1 |
| chr3_-_50277959 | 0.75 |
ENSDART00000082773
ENSDART00000139524 |
arl16
|
ADP-ribosylation factor-like 16 |
| chr24_-_33703504 | 0.75 |
ENSDART00000079292
|
cavin4b
|
caveolae associated protein 4b |
| chr17_-_31659670 | 0.75 |
ENSDART00000030448
|
vsx2
|
visual system homeobox 2 |
| chr7_+_44445595 | 0.75 |
ENSDART00000108766
|
cdh5
|
cadherin 5 |
| chr16_+_9762261 | 0.75 |
ENSDART00000020654
|
psmd4b
|
proteasome 26S subunit, non-ATPase 4b |
| chr4_+_19535946 | 0.74 |
ENSDART00000192342
ENSDART00000183740 ENSDART00000180812 ENSDART00000180017 |
lrrc4.1
|
leucine rich repeat containing 4.1 |
| chr13_+_28819768 | 0.74 |
ENSDART00000191401
ENSDART00000188895 ENSDART00000101653 |
CU639469.1
|
|
| chr2_-_30055432 | 0.74 |
ENSDART00000056747
|
shhb
|
sonic hedgehog b |
| chr23_-_3409140 | 0.74 |
ENSDART00000002309
|
mafba
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Ba |
| chr5_+_67390115 | 0.74 |
ENSDART00000193255
|
ebf2
|
early B cell factor 2 |
| chr10_+_21701568 | 0.74 |
ENSDART00000090748
|
pcdh1g9
|
protocadherin 1 gamma 9 |
| chr9_+_13985567 | 0.74 |
ENSDART00000102296
|
cd28
|
CD28 molecule |
| chr8_+_22931427 | 0.73 |
ENSDART00000063096
|
sypa
|
synaptophysin a |
| chr16_-_6377672 | 0.73 |
ENSDART00000081649
|
scgn
|
secretagogin, EF-hand calcium binding protein |
| chr21_-_22928214 | 0.73 |
ENSDART00000182760
|
dub
|
duboraya |
| chr12_-_31103906 | 0.73 |
ENSDART00000189099
ENSDART00000121527 ENSDART00000185635 ENSDART00000183754 |
tcf7l2
|
transcription factor 7 like 2 |
| chr19_+_8973042 | 0.73 |
ENSDART00000039597
|
crabp2b
|
cellular retinoic acid binding protein 2, b |
| chr21_-_32487061 | 0.72 |
ENSDART00000114359
ENSDART00000131591 ENSDART00000131477 |
si:dkeyp-72g9.4
|
si:dkeyp-72g9.4 |
| chr22_-_17606575 | 0.72 |
ENSDART00000183951
|
gpx4a
|
glutathione peroxidase 4a |
| chr22_-_3914162 | 0.72 |
ENSDART00000187174
ENSDART00000190612 ENSDART00000187928 ENSDART00000057224 ENSDART00000184758 |
mhc1uma
|
major histocompatibility complex class I UMA |
Network of associatons between targets according to the STRING database.
First level regulatory network of nfatc2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.5 | 1.0 | GO:2000257 | regulation of protein activation cascade(GO:2000257) |
| 0.5 | 1.9 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.5 | 1.4 | GO:0097378 | interneuron axon guidance(GO:0097376) spinal cord interneuron axon guidance(GO:0097377) dorsal spinal cord interneuron axon guidance(GO:0097378) |
| 0.4 | 1.3 | GO:0006824 | cobalt ion transport(GO:0006824) cobalamin transport(GO:0015889) |
| 0.4 | 1.6 | GO:0099509 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 0.4 | 1.1 | GO:0003249 | cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) endocardial cell development(GO:0060958) cell proliferation involved in heart valve development(GO:2000793) |
| 0.3 | 1.7 | GO:0010872 | regulation of cholesterol esterification(GO:0010872) positive regulation of cholesterol esterification(GO:0010873) |
| 0.3 | 1.0 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.3 | 1.3 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.3 | 2.2 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.3 | 1.8 | GO:0021767 | mammillary body development(GO:0021767) |
| 0.3 | 0.9 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) |
| 0.3 | 0.9 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.3 | 1.9 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.3 | 0.8 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.3 | 1.6 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.3 | 1.0 | GO:0006598 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.3 | 1.0 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.2 | 1.0 | GO:0060956 | cardiac endothelial cell differentiation(GO:0003348) endocardial cell differentiation(GO:0060956) |
| 0.2 | 0.7 | GO:0072020 | proximal straight tubule development(GO:0072020) |
| 0.2 | 0.7 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) |
| 0.2 | 0.9 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.2 | 0.7 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.2 | 0.7 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.2 | 1.6 | GO:0003311 | pancreatic D cell differentiation(GO:0003311) pancreatic epsilon cell differentiation(GO:0090104) |
| 0.2 | 0.6 | GO:0033335 | anal fin development(GO:0033335) |
| 0.2 | 0.6 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.2 | 0.9 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) |
| 0.2 | 0.6 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.2 | 1.2 | GO:0045063 | neutrophil apoptotic process(GO:0001781) regulation of T-helper 1 type immune response(GO:0002825) positive regulation of T-helper 1 type immune response(GO:0002827) negative regulation of type 2 immune response(GO:0002829) inflammatory cell apoptotic process(GO:0006925) regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of phospholipase A2 activity(GO:0032429) interleukin-2 production(GO:0032623) regulation of interleukin-2 production(GO:0032663) positive regulation of interleukin-2 production(GO:0032743) myeloid cell apoptotic process(GO:0033028) regulation of neutrophil apoptotic process(GO:0033029) positive regulation of neutrophil apoptotic process(GO:0033031) regulation of myeloid cell apoptotic process(GO:0033032) positive regulation of myeloid cell apoptotic process(GO:0033034) T-helper 1 type immune response(GO:0042088) positive regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043372) T-helper 1 cell differentiation(GO:0045063) positive regulation of T-helper cell differentiation(GO:0045624) regulation of T-helper 1 cell differentiation(GO:0045625) positive regulation of T-helper 1 cell differentiation(GO:0045627) regulation of T-helper 2 cell differentiation(GO:0045628) negative regulation of T-helper 2 cell differentiation(GO:0045629) positive regulation of fatty acid biosynthetic process(GO:0045723) positive regulation of alpha-beta T cell differentiation(GO:0046638) neutrophil clearance(GO:0097350) negative regulation of phospholipase A2 activity(GO:1900138) positive regulation of CD4-positive, alpha-beta T cell activation(GO:2000516) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.2 | 0.6 | GO:0070221 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.2 | 0.8 | GO:0003418 | chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 0.2 | 0.8 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.2 | 0.9 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.2 | 0.7 | GO:0055107 | Golgi to secretory granule transport(GO:0055107) |
| 0.2 | 1.1 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.2 | 0.7 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.2 | 0.7 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.2 | 1.0 | GO:0010801 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.2 | 0.5 | GO:0002432 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 0.2 | 1.0 | GO:0055016 | hypochord development(GO:0055016) |
| 0.2 | 0.8 | GO:0046457 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.2 | 0.7 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.2 | 0.5 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.2 | 0.7 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.2 | 1.8 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.2 | 1.0 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.2 | 1.6 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.2 | 2.8 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.1 | 0.7 | GO:0033292 | T-tubule organization(GO:0033292) |
| 0.1 | 2.4 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.1 | 2.0 | GO:0048385 | regulation of retinoic acid receptor signaling pathway(GO:0048385) |
| 0.1 | 0.7 | GO:0071908 | determination of intestine left/right asymmetry(GO:0071908) |
| 0.1 | 0.1 | GO:0007440 | foregut morphogenesis(GO:0007440) post-embryonic foregut morphogenesis(GO:0048618) |
| 0.1 | 1.0 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.1 | 1.0 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.1 | 0.7 | GO:0034653 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.1 | 0.8 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.1 | 0.4 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.1 | 0.9 | GO:0030194 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.1 | 0.9 | GO:0060855 | venous endothelial cell migration involved in lymph vessel development(GO:0060855) |
| 0.1 | 0.7 | GO:0006154 | adenosine catabolic process(GO:0006154) inosine metabolic process(GO:0046102) inosine biosynthetic process(GO:0046103) |
| 0.1 | 0.5 | GO:0015868 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.1 | 0.5 | GO:0060547 | negative regulation of necroptotic process(GO:0060546) negative regulation of necrotic cell death(GO:0060547) |
| 0.1 | 0.6 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.1 | 0.2 | GO:0045429 | regulation of nitric oxide biosynthetic process(GO:0045428) positive regulation of nitric oxide biosynthetic process(GO:0045429) positive regulation of reactive oxygen species biosynthetic process(GO:1903428) positive regulation of nitric oxide metabolic process(GO:1904407) |
| 0.1 | 1.6 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.1 | 1.8 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) |
| 0.1 | 0.4 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.1 | 1.1 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.1 | 1.1 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 | 0.4 | GO:0071498 | cellular response to fluid shear stress(GO:0071498) |
| 0.1 | 0.5 | GO:0043011 | myeloid dendritic cell activation(GO:0001773) myeloid dendritic cell differentiation(GO:0043011) |
| 0.1 | 0.8 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 0.3 | GO:1901890 | positive regulation of cell junction assembly(GO:1901890) |
| 0.1 | 0.8 | GO:0090280 | positive regulation of calcium ion import(GO:0090280) |
| 0.1 | 0.2 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.1 | 2.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 0.4 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.1 | 0.7 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 | 0.5 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 2.3 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 0.8 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.1 | 1.1 | GO:0019233 | sensory perception of pain(GO:0019233) |
| 0.1 | 0.8 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.1 | 1.3 | GO:0003209 | cardiac atrium morphogenesis(GO:0003209) |
| 0.1 | 0.2 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) |
| 0.1 | 0.7 | GO:0071678 | olfactory bulb axon guidance(GO:0071678) |
| 0.1 | 0.8 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.1 | 0.6 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 0.1 | 0.3 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 0.4 | GO:0090299 | regulation of neural crest formation(GO:0090299) |
| 0.1 | 0.9 | GO:0014028 | notochord formation(GO:0014028) |
| 0.1 | 0.4 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.1 | 0.4 | GO:0021982 | pineal gland development(GO:0021982) |
| 0.1 | 0.9 | GO:0032099 | negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.1 | 0.3 | GO:0045577 | regulation of B cell differentiation(GO:0045577) |
| 0.1 | 0.3 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.1 | 0.3 | GO:0010934 | macrophage cytokine production(GO:0010934) regulation of macrophage cytokine production(GO:0010935) positive regulation of macrophage cytokine production(GO:0060907) |
| 0.1 | 2.4 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.1 | 0.7 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.1 | 0.2 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.1 | 0.6 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 0.4 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.1 | 1.6 | GO:0042159 | lipoprotein catabolic process(GO:0042159) |
| 0.1 | 2.5 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.1 | 0.4 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.2 | GO:1903173 | fatty alcohol metabolic process(GO:1903173) |
| 0.1 | 0.9 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 0.4 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.1 | 0.3 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.2 | GO:1901232 | regulation of convergent extension involved in axis elongation(GO:1901232) positive regulation of epiboly involved in gastrulation with mouth forming second(GO:1904088) |
| 0.1 | 0.3 | GO:0014909 | smooth muscle cell migration(GO:0014909) |
| 0.1 | 0.4 | GO:0070973 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 1.4 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.1 | 0.3 | GO:1900120 | microglial cell activation(GO:0001774) regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 0.1 | 0.7 | GO:0060114 | vestibular receptor cell differentiation(GO:0060114) vestibular receptor cell development(GO:0060118) |
| 0.1 | 1.1 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.1 | 0.3 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.1 | 0.4 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.1 | 0.2 | GO:0048795 | swim bladder morphogenesis(GO:0048795) |
| 0.1 | 0.4 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.1 | 0.8 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.1 | 0.1 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.1 | 1.1 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.1 | 0.7 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.1 | 0.5 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.1 | 0.4 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 | 0.5 | GO:0090497 | mesenchymal cell migration(GO:0090497) |
| 0.1 | 0.6 | GO:2001046 | positive regulation of integrin-mediated signaling pathway(GO:2001046) |
| 0.1 | 0.3 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.1 | 0.3 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.1 | 0.3 | GO:1903385 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 0.3 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.1 | 0.7 | GO:0030656 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.1 | 0.2 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.1 | 0.5 | GO:0097531 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.1 | 0.6 | GO:0018120 | peptidyl-arginine ADP-ribosylation(GO:0018120) |
| 0.1 | 1.2 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.1 | 0.3 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 0.2 | GO:0060437 | lung growth(GO:0060437) secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.1 | 0.1 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.1 | 1.0 | GO:0033077 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.1 | 0.2 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.1 | 0.5 | GO:0048796 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.1 | 0.3 | GO:2000301 | negative regulation of neurotransmitter secretion(GO:0046929) response to cAMP(GO:0051591) cellular response to cAMP(GO:0071320) negative regulation of synaptic vesicle transport(GO:1902804) negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.1 | 0.6 | GO:1902667 | regulation of axon guidance(GO:1902667) |
| 0.1 | 0.6 | GO:0015800 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.1 | 1.2 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.1 | 1.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 0.2 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 | 0.1 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.1 | 0.2 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.1 | 0.3 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.1 | 0.2 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 0.2 | GO:0097052 | L-kynurenine metabolic process(GO:0097052) L-kynurenine catabolic process(GO:0097053) |
| 0.1 | 1.2 | GO:0048512 | rhythmic behavior(GO:0007622) circadian behavior(GO:0048512) |
| 0.1 | 1.8 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.1 | 1.7 | GO:0021761 | limbic system development(GO:0021761) |
| 0.1 | 0.5 | GO:0043097 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) pyrimidine nucleoside salvage(GO:0043097) UMP salvage(GO:0044206) |
| 0.1 | 1.6 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.1 | 0.7 | GO:0060034 | notochord cell differentiation(GO:0060034) |
| 0.1 | 1.5 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.1 | 0.5 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 0.1 | GO:0009097 | isoleucine metabolic process(GO:0006549) isoleucine biosynthetic process(GO:0009097) |
| 0.1 | 0.4 | GO:0030324 | respiratory tube development(GO:0030323) lung development(GO:0030324) |
| 0.1 | 0.3 | GO:0021576 | hindbrain formation(GO:0021576) |
| 0.0 | 0.3 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.0 | 0.2 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.7 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.2 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.0 | 0.5 | GO:0097090 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.4 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.0 | 0.2 | GO:0009791 | post-embryonic development(GO:0009791) |
| 0.0 | 2.8 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 0.4 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 2.2 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.1 | GO:0042941 | D-amino acid transport(GO:0042940) D-alanine transport(GO:0042941) D-serine transport(GO:0042942) |
| 0.0 | 0.2 | GO:0051204 | protein insertion into membrane from inner side(GO:0032978) protein insertion into mitochondrial membrane from inner side(GO:0032979) protein insertion into mitochondrial membrane(GO:0051204) |
| 0.0 | 4.7 | GO:0033333 | fin development(GO:0033333) |
| 0.0 | 1.8 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.9 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.6 | GO:0035860 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.9 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.0 | 1.2 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.1 | GO:0046087 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.4 | GO:0021984 | adenohypophysis development(GO:0021984) |
| 0.0 | 0.3 | GO:0098971 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.0 | 0.2 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.8 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.0 | 0.1 | GO:1903673 | mitotic cytokinetic process(GO:1902410) mitotic cleavage furrow formation(GO:1903673) |
| 0.0 | 0.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.5 | GO:0030809 | negative regulation of adenylate cyclase activity(GO:0007194) negative regulation of cyclic nucleotide metabolic process(GO:0030800) negative regulation of cyclic nucleotide biosynthetic process(GO:0030803) negative regulation of nucleotide biosynthetic process(GO:0030809) negative regulation of cAMP metabolic process(GO:0030815) negative regulation of cAMP biosynthetic process(GO:0030818) negative regulation of cyclase activity(GO:0031280) negative regulation of lyase activity(GO:0051350) negative regulation of purine nucleotide biosynthetic process(GO:1900372) |
| 0.0 | 0.4 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.3 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.0 | 0.2 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.0 | 0.3 | GO:0090594 | inflammatory response to wounding(GO:0090594) |
| 0.0 | 0.8 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.2 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.0 | 1.3 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 | 0.3 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.3 | GO:0071679 | commissural neuron axon guidance(GO:0071679) |
| 0.0 | 0.2 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 1.2 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 0.5 | GO:0007140 | male meiosis(GO:0007140) |
| 0.0 | 0.7 | GO:0048679 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.0 | 0.2 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 7.1 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 | 0.6 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.2 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.3 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.1 | GO:0034695 | response to prostaglandin E(GO:0034695) cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.0 | 0.2 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.0 | 0.3 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.0 | 0.1 | GO:0050764 | regulation of phagocytosis(GO:0050764) regulation of apoptotic cell clearance(GO:2000425) |
| 0.0 | 0.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.3 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.9 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.3 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.5 | GO:0045807 | positive regulation of endocytosis(GO:0045807) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.3 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.4 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.1 | GO:0071387 | response to cortisol(GO:0051414) cellular response to cortisol stimulus(GO:0071387) |
| 0.0 | 0.3 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.3 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.6 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.2 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.5 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.7 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.7 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.4 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.3 | GO:0060396 | growth hormone receptor signaling pathway(GO:0060396) response to growth hormone(GO:0060416) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.6 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.5 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.3 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.6 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 1.0 | GO:0044259 | collagen metabolic process(GO:0032963) multicellular organismal macromolecule metabolic process(GO:0044259) |
| 0.0 | 0.2 | GO:0043090 | amino acid import(GO:0043090) L-arginine import(GO:0043091) L-amino acid import(GO:0043092) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.1 | GO:0034331 | cell junction maintenance(GO:0034331) |
| 0.0 | 0.4 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 0.5 | GO:0006183 | GTP biosynthetic process(GO:0006183) |
| 0.0 | 0.2 | GO:0043153 | entrainment of circadian clock(GO:0009649) entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.3 | GO:1902042 | regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902041) negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.0 | 0.7 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 1.1 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.5 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.0 | 16.5 | GO:0006955 | immune response(GO:0006955) |
| 0.0 | 1.4 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.4 | GO:0048678 | axon regeneration(GO:0031103) response to axon injury(GO:0048678) |
| 0.0 | 0.6 | GO:0001649 | osteoblast differentiation(GO:0001649) |
| 0.0 | 3.9 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 0.2 | GO:1900407 | regulation of cellular response to oxidative stress(GO:1900407) |
| 0.0 | 0.8 | GO:0021515 | cell differentiation in spinal cord(GO:0021515) |
| 0.0 | 0.2 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.6 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 5.6 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.4 | GO:0001819 | positive regulation of cytokine production(GO:0001819) |
| 0.0 | 0.1 | GO:0050688 | regulation of response to biotic stimulus(GO:0002831) regulation of defense response to virus(GO:0050688) |
| 0.0 | 0.2 | GO:0000050 | urea cycle(GO:0000050) urea metabolic process(GO:0019627) |
| 0.0 | 0.1 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) nucleobase catabolic process(GO:0046113) |
| 0.0 | 0.2 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.0 | 3.0 | GO:1990138 | neuron projection extension(GO:1990138) |
| 0.0 | 0.2 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.3 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.0 | 1.0 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.1 | GO:0010359 | regulation of anion channel activity(GO:0010359) |
| 0.0 | 0.1 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 0.3 | GO:0072102 | glomerulus morphogenesis(GO:0072102) |
| 0.0 | 1.2 | GO:1904888 | cranial skeletal system development(GO:1904888) |
| 0.0 | 0.1 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 | 0.2 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.6 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.9 | GO:0007599 | hemostasis(GO:0007599) |
| 0.0 | 0.1 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 1.2 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.0 | 0.5 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.4 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0045852 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.0 | 1.7 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 0.1 | GO:0070376 | ERK5 cascade(GO:0070375) regulation of ERK5 cascade(GO:0070376) positive regulation of ERK5 cascade(GO:0070378) |
| 0.0 | 0.1 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.0 | 0.2 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.0 | 0.4 | GO:1902622 | regulation of neutrophil migration(GO:1902622) |
| 0.0 | 0.1 | GO:0009182 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) deoxyribonucleoside triphosphate biosynthetic process(GO:0009202) dGDP metabolic process(GO:0046066) |
| 0.0 | 0.1 | GO:0045665 | negative regulation of neuron differentiation(GO:0045665) |
| 0.0 | 0.9 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.0 | 0.5 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.0 | 0.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.5 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.4 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.4 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.1 | GO:0016121 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.0 | 0.6 | GO:0048840 | otolith development(GO:0048840) |
| 0.0 | 0.3 | GO:0070897 | DNA-templated transcriptional preinitiation complex assembly(GO:0070897) |
| 0.0 | 0.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.2 | GO:0044247 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.1 | GO:0021654 | rhombomere boundary formation(GO:0021654) |
| 0.0 | 0.4 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 1.3 | GO:1901342 | regulation of vasculature development(GO:1901342) |
| 0.0 | 1.0 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.2 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
| 0.0 | 0.5 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.2 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.8 | GO:0002521 | leukocyte differentiation(GO:0002521) |
| 0.0 | 0.3 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.1 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.0 | 0.4 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 0.0 | GO:1990416 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 0.4 | GO:2001056 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043280) positive regulation of cysteine-type endopeptidase activity(GO:2001056) |
| 0.0 | 0.1 | GO:0002183 | cytoplasmic translational initiation(GO:0002183) |
| 0.0 | 0.3 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.3 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 0.5 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.5 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.9 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.3 | 2.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.3 | 3.4 | GO:0042627 | chylomicron(GO:0042627) |
| 0.2 | 1.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.2 | 1.5 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.2 | 1.8 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 0.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.4 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.1 | 2.6 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 1.3 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.1 | 1.8 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 1.0 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 1.4 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 0.4 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.1 | 0.3 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 0.3 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.1 | 1.7 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.1 | 0.5 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 6.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 0.4 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.1 | 0.7 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 1.3 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 0.5 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 0.5 | GO:0071914 | prominosome(GO:0071914) |
| 0.1 | 2.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.2 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.1 | 0.2 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.1 | 15.8 | GO:0005764 | lysosome(GO:0005764) |
| 0.1 | 0.9 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 2.0 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 0.9 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 0.7 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 1.0 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.8 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 1.0 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.3 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.0 | 1.7 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.5 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 2.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 2.5 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.0 | 0.6 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.3 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.1 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.0 | 0.4 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 1.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.3 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.1 | GO:0005880 | nuclear microtubule(GO:0005880) |
| 0.0 | 0.2 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 1.5 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.8 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 3.2 | GO:0031674 | I band(GO:0031674) |
| 0.0 | 0.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.5 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.6 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 8.9 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.1 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.3 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 24.7 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.4 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0005913 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.0 | 1.4 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 0.2 | GO:0030017 | sarcomere(GO:0030017) |
| 0.0 | 0.4 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.3 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 1.1 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.8 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 10.8 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 0.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.4 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.7 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.1 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.7 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.3 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.0 | 0.0 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.5 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.8 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.8 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 1.1 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.0 | 0.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 1.3 | GO:0005925 | cell-substrate adherens junction(GO:0005924) focal adhesion(GO:0005925) |
| 0.0 | 1.3 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.0 | 0.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 1.0 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.1 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.2 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.1 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.9 | GO:0008430 | selenium binding(GO:0008430) |
| 0.3 | 2.8 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.3 | 1.7 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.3 | 1.2 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.3 | 0.9 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.3 | 1.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.2 | 1.0 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.2 | 0.7 | GO:0009013 | succinate-semialdehyde dehydrogenase (NAD+) activity(GO:0004777) succinate-semialdehyde dehydrogenase [NAD(P)+] activity(GO:0009013) |
| 0.2 | 0.9 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.2 | 0.7 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.2 | 0.6 | GO:0008457 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.2 | 0.8 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.2 | 5.8 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.2 | 1.2 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.2 | 0.6 | GO:0070224 | sulfide:quinone oxidoreductase activity(GO:0070224) |
| 0.2 | 0.6 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.2 | 0.8 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.2 | 0.7 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.2 | 0.8 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.2 | 0.6 | GO:0031544 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.2 | 0.5 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.2 | 1.3 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.2 | 0.5 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.2 | 1.8 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.1 | 1.3 | GO:0005113 | patched binding(GO:0005113) |
| 0.1 | 1.0 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.1 | 0.7 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.1 | 0.6 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.1 | 0.4 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.1 | 21.1 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 0.7 | GO:0016519 | gastric inhibitory peptide receptor activity(GO:0016519) |
| 0.1 | 0.4 | GO:0043185 | vascular endothelial growth factor receptor 3 binding(GO:0043185) |
| 0.1 | 0.5 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 0.5 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.1 | 0.5 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.1 | 0.8 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.1 | 0.5 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 0.5 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.1 | 0.9 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 1.6 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 1.0 | GO:0005402 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.1 | 1.1 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 1.6 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.1 | 0.5 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.1 | 0.5 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 1.4 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 2.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.9 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 0.2 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.1 | 0.4 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.7 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 0.4 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.1 | 1.4 | GO:0043236 | laminin binding(GO:0043236) |
| 0.1 | 1.4 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 0.5 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.1 | 0.4 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.1 | 0.5 | GO:0019863 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.1 | 0.4 | GO:0031781 | type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.1 | 0.6 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.1 | 1.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 5.3 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 0.4 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 0.3 | GO:0048407 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 1.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 0.3 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.1 | 0.3 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 0.6 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.1 | 1.1 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.1 | 8.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 0.5 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.1 | 0.5 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 1.4 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.1 | 0.9 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 0.6 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.1 | 0.4 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.1 | GO:0038046 | enkephalin receptor activity(GO:0038046) |
| 0.1 | 0.2 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.1 | 0.5 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 1.2 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 0.5 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.1 | 0.3 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.1 | 0.9 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.1 | 0.2 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.1 | 0.6 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 0.4 | GO:0005035 | death receptor activity(GO:0005035) |
| 0.1 | 0.3 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 0.5 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 0.3 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.1 | 0.3 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 0.3 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 0.5 | GO:0019825 | oxygen transporter activity(GO:0005344) oxygen binding(GO:0019825) |
| 0.0 | 1.0 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 1.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.6 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.2 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) S-(hydroxymethyl)glutathione dehydrogenase activity(GO:0051903) |
| 0.0 | 0.1 | GO:0003999 | adenine phosphoribosyltransferase activity(GO:0003999) |
| 0.0 | 0.2 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.0 | 0.9 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.3 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.0 | 0.5 | GO:0051373 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.0 | 0.2 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.0 | 0.2 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.0 | 0.9 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 1.2 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.4 | GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) |
| 0.0 | 0.3 | GO:0004960 | thromboxane receptor activity(GO:0004960) |
| 0.0 | 1.1 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 7.0 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.9 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.2 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 0.9 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0031530 | gonadotropin hormone-releasing hormone activity(GO:0005183) gonadotropin-releasing hormone receptor binding(GO:0031530) |
| 0.0 | 0.5 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 0.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.7 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.3 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.6 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.2 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.5 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.3 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.6 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.4 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.3 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.3 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.1 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) type II activin receptor binding(GO:0070699) |
| 0.0 | 0.5 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 1.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0071568 | UFM1 transferase activity(GO:0071568) |
| 0.0 | 0.4 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.0 | 0.5 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.3 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.4 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 2.4 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.9 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.4 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.4 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.4 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 1.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.4 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.2 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 1.0 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.5 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.0 | 0.2 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 4.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.2 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.5 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 1.5 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.3 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.5 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 1.1 | GO:0005539 | glycosaminoglycan binding(GO:0005539) |
| 0.0 | 0.2 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 3.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 20.3 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.5 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.1 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.5 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.9 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.4 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.3 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.1 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) |
| 0.0 | 0.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.2 | GO:0010436 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.0 | 0.1 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.0 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.0 | 0.3 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.2 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.4 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.3 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 1.1 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.1 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.2 | GO:0030546 | receptor activator activity(GO:0030546) receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 23.3 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.2 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.8 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.2 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.3 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.4 | GO:0005343 | organic acid:sodium symporter activity(GO:0005343) |
| 0.0 | 0.1 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.0 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.0 | 0.2 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.4 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.1 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 1.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 1.8 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 0.5 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.1 | 1.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 0.4 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 0.8 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 1.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 1.2 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 0.8 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 2.5 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 0.4 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.1 | 2.5 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 7.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 0.8 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.7 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.2 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 3.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.3 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.4 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.5 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.2 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 5.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.8 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.6 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.3 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.5 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.4 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.3 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 1.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.6 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.3 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.3 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.4 | NABA CORE MATRISOME | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans |
| 0.0 | 0.2 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.1 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.3 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.6 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.2 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.2 | 1.8 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 0.4 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.1 | 0.9 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 1.2 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.1 | 0.4 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.1 | 0.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 0.4 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 0.8 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 0.3 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.1 | 0.4 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 0.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 0.8 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 1.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 0.8 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.1 | 0.4 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 2.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 1.4 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.1 | 1.3 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.1 | 0.3 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 0.7 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 0.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.4 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.2 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.5 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 1.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 3.9 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.5 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 1.4 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 1.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 0.5 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.0 | 1.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.5 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 1.0 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.2 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.3 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.3 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.5 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.2 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 1.3 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.3 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.1 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.5 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.6 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.1 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.4 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.1 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |