Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
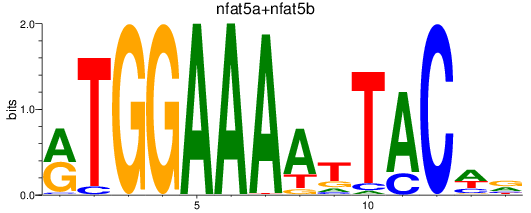
Results for nfat5a+nfat5b
Z-value: 0.49
Transcription factors associated with nfat5a+nfat5b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nfat5a
|
ENSDARG00000020872 | nuclear factor of activated T cells 5a |
|
nfat5b
|
ENSDARG00000102556 | nuclear factor of activated T cells 5b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nfat5a | dr11_v1_chr25_-_37121335_37121335 | -0.43 | 2.5e-01 | Click! |
| nfat5b | dr11_v1_chr7_+_67325933_67325933 | -0.36 | 3.4e-01 | Click! |
Activity profile of nfat5a+nfat5b motif
Sorted Z-values of nfat5a+nfat5b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_39444843 | 0.70 |
ENSDART00000143999
ENSDART00000173554 ENSDART00000173698 ENSDART00000173754 ENSDART00000144075 ENSDART00000138192 ENSDART00000145457 ENSDART00000141750 ENSDART00000103056 ENSDART00000142946 ENSDART00000173748 |
tnnt3b
|
troponin T type 3b (skeletal, fast) |
| chr16_+_23398369 | 0.69 |
ENSDART00000037694
|
s100a10b
|
S100 calcium binding protein A10b |
| chr16_+_23397785 | 0.67 |
ENSDART00000148961
|
s100a10b
|
S100 calcium binding protein A10b |
| chr2_+_55982940 | 0.59 |
ENSDART00000097753
ENSDART00000097751 |
nmrk2
|
nicotinamide riboside kinase 2 |
| chr9_+_53725294 | 0.57 |
ENSDART00000165991
|
cnmd
|
chondromodulin |
| chr9_+_53725128 | 0.55 |
ENSDART00000169062
|
cnmd
|
chondromodulin |
| chr15_+_33989181 | 0.53 |
ENSDART00000169487
|
vwde
|
von Willebrand factor D and EGF domains |
| chr2_+_55982300 | 0.50 |
ENSDART00000183903
|
nmrk2
|
nicotinamide riboside kinase 2 |
| chr6_+_2097690 | 0.38 |
ENSDART00000193770
|
tgm2b
|
transglutaminase 2b |
| chr16_+_31804590 | 0.37 |
ENSDART00000167321
|
wnt4b
|
wingless-type MMTV integration site family, member 4b |
| chr24_-_24060632 | 0.35 |
ENSDART00000090514
|
abcc13
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 13 |
| chr19_-_8748571 | 0.29 |
ENSDART00000031173
|
rps27.1
|
ribosomal protein S27, isoform 1 |
| chr1_-_26293203 | 0.29 |
ENSDART00000180140
|
cxxc4
|
CXXC finger 4 |
| chr1_+_19303241 | 0.28 |
ENSDART00000129970
|
si:dkeyp-118a3.2
|
si:dkeyp-118a3.2 |
| chr9_+_310331 | 0.28 |
ENSDART00000172446
ENSDART00000187731 ENSDART00000193970 |
stac3
|
SH3 and cysteine rich domain 3 |
| chr16_-_28836523 | 0.27 |
ENSDART00000108596
ENSDART00000142920 |
plin6
|
perilipin 6 |
| chr14_-_25599002 | 0.27 |
ENSDART00000040955
|
slc25a48
|
solute carrier family 25, member 48 |
| chr9_+_33145522 | 0.26 |
ENSDART00000005879
|
atp5po
|
ATP synthase peripheral stalk subunit OSCP |
| chr23_+_36771593 | 0.26 |
ENSDART00000078240
|
march9
|
membrane-associated ring finger (C3HC4) 9 |
| chr20_-_40720458 | 0.26 |
ENSDART00000153151
ENSDART00000061261 ENSDART00000138569 |
cx43
|
connexin 43 |
| chr20_-_29787192 | 0.26 |
ENSDART00000125348
ENSDART00000048759 |
id2b
|
inhibitor of DNA binding 2b |
| chr5_+_67971627 | 0.26 |
ENSDART00000144879
|
mtif3
|
mitochondrial translational initiation factor 3 |
| chr8_+_31872992 | 0.25 |
ENSDART00000146933
|
c7a
|
complement component 7a |
| chr2_-_6115688 | 0.24 |
ENSDART00000081663
|
prdx1
|
peroxiredoxin 1 |
| chr24_-_21923930 | 0.24 |
ENSDART00000131944
|
tagln3b
|
transgelin 3b |
| chr7_-_48667056 | 0.24 |
ENSDART00000006378
|
cdkn1ca
|
cyclin-dependent kinase inhibitor 1Ca |
| chr25_+_34013093 | 0.22 |
ENSDART00000011967
|
anxa2a
|
annexin A2a |
| chr16_+_20895904 | 0.22 |
ENSDART00000052662
|
hoxa13b
|
homeobox A13b |
| chr10_+_22782522 | 0.22 |
ENSDART00000079498
ENSDART00000145558 |
si:ch211-237l4.6
|
si:ch211-237l4.6 |
| chr1_-_26292897 | 0.22 |
ENSDART00000112899
ENSDART00000185410 |
cxxc4
|
CXXC finger 4 |
| chr2_-_41518340 | 0.21 |
ENSDART00000130830
|
otomp
|
otolith matrix protein |
| chr12_-_30549022 | 0.21 |
ENSDART00000102474
|
zgc:158404
|
zgc:158404 |
| chr18_-_14860435 | 0.20 |
ENSDART00000018502
|
mapk12a
|
mitogen-activated protein kinase 12a |
| chr10_+_13209580 | 0.20 |
ENSDART00000000887
ENSDART00000136932 |
rassf6
|
Ras association (RalGDS/AF-6) domain family 6 |
| chr10_+_21730585 | 0.20 |
ENSDART00000188576
|
pcdh1g22
|
protocadherin 1 gamma 22 |
| chr6_+_49095646 | 0.19 |
ENSDART00000103385
|
slc25a55a
|
solute carrier family 25, member 55a |
| chr23_+_36122058 | 0.18 |
ENSDART00000184448
|
hoxc3a
|
homeobox C3a |
| chr24_-_21674950 | 0.18 |
ENSDART00000123216
ENSDART00000046211 |
lnx2a
|
ligand of numb-protein X 2a |
| chr20_+_34326874 | 0.18 |
ENSDART00000061659
|
ivns1abpa
|
influenza virus NS1A binding protein a |
| chr21_+_26657404 | 0.18 |
ENSDART00000129035
ENSDART00000186550 |
prdx5
|
peroxiredoxin 5 |
| chr2_+_31382590 | 0.18 |
ENSDART00000086863
|
colec12
|
collectin sub-family member 12 |
| chr23_+_28770225 | 0.17 |
ENSDART00000132179
ENSDART00000142273 |
masp2
|
mannan-binding lectin serine peptidase 2 |
| chr15_+_1796313 | 0.17 |
ENSDART00000126253
|
fam124b
|
family with sequence similarity 124B |
| chr13_-_42749916 | 0.17 |
ENSDART00000140019
|
capn2a
|
calpain 2, (m/II) large subunit a |
| chr5_-_37900350 | 0.16 |
ENSDART00000084839
ENSDART00000084841 ENSDART00000133437 |
tmprss13b
|
transmembrane protease, serine 13b |
| chr3_-_31019715 | 0.15 |
ENSDART00000156687
ENSDART00000156127 ENSDART00000153945 |
si:dkeyp-71f10.5
|
si:dkeyp-71f10.5 |
| chr3_-_32590164 | 0.15 |
ENSDART00000151151
|
tspan4b
|
tetraspanin 4b |
| chr18_-_898870 | 0.15 |
ENSDART00000151777
ENSDART00000062654 |
parp6a
|
poly (ADP-ribose) polymerase family, member 6a |
| chr21_+_5494561 | 0.14 |
ENSDART00000169728
|
ly6m3
|
lymphocyte antigen 6 family member M3 |
| chr13_-_11667661 | 0.14 |
ENSDART00000102411
|
dctn1b
|
dynactin 1b |
| chr5_-_12713920 | 0.13 |
ENSDART00000099749
|
CLDN22
|
si:dkey-98f17.3 |
| chr2_-_43151358 | 0.13 |
ENSDART00000061170
ENSDART00000056161 |
ftr15
|
finTRIM family, member 15 |
| chr4_+_76722754 | 0.13 |
ENSDART00000153867
|
ms4a17a.3
|
membrane-spanning 4-domains, subfamily A, member 17A.3 |
| chr3_-_35554809 | 0.13 |
ENSDART00000010944
|
dctn5
|
dynactin 5 |
| chr1_-_22834824 | 0.12 |
ENSDART00000043556
|
ldb2b
|
LIM domain binding 2b |
| chr25_+_7461624 | 0.12 |
ENSDART00000170569
|
syt12
|
synaptotagmin XII |
| chr2_-_20923864 | 0.12 |
ENSDART00000006870
|
ptgs2a
|
prostaglandin-endoperoxide synthase 2a |
| chr22_-_32360464 | 0.12 |
ENSDART00000148886
|
pcbp4
|
poly(rC) binding protein 4 |
| chr6_-_57635577 | 0.11 |
ENSDART00000168708
|
cbfa2t2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2 |
| chr10_+_21722892 | 0.11 |
ENSDART00000162855
|
pcdh1g13
|
protocadherin 1 gamma 13 |
| chr11_-_12051805 | 0.11 |
ENSDART00000110117
ENSDART00000182744 |
socs7
|
suppressor of cytokine signaling 7 |
| chr7_-_56793739 | 0.11 |
ENSDART00000082842
|
si:ch211-146m13.3
|
si:ch211-146m13.3 |
| chr21_+_22840246 | 0.10 |
ENSDART00000151621
|
birc2
|
baculoviral IAP repeat containing 2 |
| chr7_+_56098590 | 0.10 |
ENSDART00000098453
|
cdh15
|
cadherin 15, type 1, M-cadherin (myotubule) |
| chr23_-_26522760 | 0.10 |
ENSDART00000142417
ENSDART00000135606 ENSDART00000122668 |
si:dkey-205h13.1
|
si:dkey-205h13.1 |
| chr8_+_7144066 | 0.09 |
ENSDART00000146306
|
slc6a6a
|
solute carrier family 6 (neurotransmitter transporter), member 6a |
| chr7_-_31941670 | 0.09 |
ENSDART00000180929
ENSDART00000075389 |
bdnf
|
brain-derived neurotrophic factor |
| chr5_-_42904329 | 0.09 |
ENSDART00000112807
|
cxcl20
|
chemokine (C-X-C motif) ligand 20 |
| chr10_-_37072776 | 0.09 |
ENSDART00000110835
|
myo18aa
|
myosin XVIIIAa |
| chr24_-_24060460 | 0.08 |
ENSDART00000142813
|
abcc13
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 13 |
| chr21_+_44557006 | 0.08 |
ENSDART00000170159
|
cmc4
|
C-x(9)-C motif containing 4 homolog (S. cerevisiae) |
| chr3_+_40856095 | 0.08 |
ENSDART00000143207
|
mmd2a
|
monocyte to macrophage differentiation-associated 2a |
| chr13_-_50139916 | 0.08 |
ENSDART00000099475
|
nid1a
|
nidogen 1a |
| chr18_+_45792035 | 0.08 |
ENSDART00000135045
|
abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr3_-_29688370 | 0.08 |
ENSDART00000151147
ENSDART00000151679 |
si:ch73-233k15.2
|
si:ch73-233k15.2 |
| chr20_-_1174266 | 0.08 |
ENSDART00000023304
|
gabrr2b
|
gamma-aminobutyric acid (GABA) A receptor, rho 2b |
| chr2_-_37888429 | 0.08 |
ENSDART00000183277
|
mbl2
|
mannose binding lectin 2 |
| chr2_-_37960688 | 0.07 |
ENSDART00000055565
|
cbln14
|
cerebellin 14 |
| chr25_+_7321675 | 0.07 |
ENSDART00000104712
ENSDART00000142934 |
hmg20a
|
high mobility group 20A |
| chr25_-_13842618 | 0.07 |
ENSDART00000160258
|
mapk8ip1a
|
mitogen-activated protein kinase 8 interacting protein 1a |
| chr2_-_37862380 | 0.07 |
ENSDART00000186005
|
si:ch211-284o19.8
|
si:ch211-284o19.8 |
| chr21_-_17603182 | 0.06 |
ENSDART00000020048
ENSDART00000177270 |
gsna
|
gelsolin a |
| chr5_-_63109232 | 0.06 |
ENSDART00000115128
|
usp2b
|
ubiquitin specific peptidase 2b |
| chr7_+_72012397 | 0.06 |
ENSDART00000011303
|
ywhae2
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide 2 |
| chr6_-_42073367 | 0.06 |
ENSDART00000154304
|
grm2a
|
glutamate receptor, metabotropic 2a |
| chr7_+_57795974 | 0.06 |
ENSDART00000148369
|
camk2d1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 1 |
| chr6_-_50704689 | 0.06 |
ENSDART00000074100
|
osgn1
|
oxidative stress induced growth inhibitor 1 |
| chr14_+_23970818 | 0.06 |
ENSDART00000123338
ENSDART00000124944 |
kif3a
|
kinesin family member 3A |
| chr20_+_10498986 | 0.06 |
ENSDART00000064114
|
zgc:100997
|
zgc:100997 |
| chr5_-_49012569 | 0.06 |
ENSDART00000184690
|
si:dkey-172m14.2
|
si:dkey-172m14.2 |
| chr4_+_57093908 | 0.05 |
ENSDART00000170198
|
si:ch211-238e22.5
|
si:ch211-238e22.5 |
| chr10_-_42882215 | 0.05 |
ENSDART00000180580
ENSDART00000187374 |
FP236542.1
|
|
| chr13_-_41155472 | 0.05 |
ENSDART00000160588
|
si:dkeyp-86d6.2
|
si:dkeyp-86d6.2 |
| chr7_+_39624728 | 0.05 |
ENSDART00000173847
ENSDART00000173845 |
ptpn5
|
protein tyrosine phosphatase, non-receptor type 5 |
| chr20_+_10498704 | 0.04 |
ENSDART00000192985
|
zgc:100997
|
zgc:100997 |
| chr22_-_8306743 | 0.04 |
ENSDART00000123982
|
CABZ01077218.1
|
|
| chr16_+_29303971 | 0.04 |
ENSDART00000087149
|
hapln2
|
hyaluronan and proteoglycan link protein 2 |
| chr21_+_2742823 | 0.04 |
ENSDART00000189004
|
CABZ01073424.2
|
|
| chr16_-_21047872 | 0.04 |
ENSDART00000131582
|
cbx3b
|
chromobox homolog 3b |
| chr2_-_36925561 | 0.04 |
ENSDART00000187690
|
map1sb
|
microtubule-associated protein 1Sb |
| chr1_+_56502706 | 0.04 |
ENSDART00000188665
|
CABZ01059403.1
|
|
| chr11_-_236984 | 0.03 |
ENSDART00000170778
|
dusp7
|
dual specificity phosphatase 7 |
| chr4_+_47751483 | 0.03 |
ENSDART00000160043
|
si:ch211-196f19.1
|
si:ch211-196f19.1 |
| chr12_-_17024079 | 0.03 |
ENSDART00000129027
|
ifit11
|
interferon-induced protein with tetratricopeptide repeats 11 |
| chr15_+_37950556 | 0.03 |
ENSDART00000153947
|
si:dkey-238d18.12
|
si:dkey-238d18.12 |
| chr2_-_34483597 | 0.03 |
ENSDART00000133224
|
brinp2
|
bone morphogenetic protein/retinoic acid inducible neural-specific 2 |
| chr6_+_51932563 | 0.03 |
ENSDART00000181778
|
angpt4
|
angiopoietin 4 |
| chr17_+_24756379 | 0.03 |
ENSDART00000185059
|
stac
|
SH3 and cysteine rich domain |
| chr1_-_29658721 | 0.03 |
ENSDART00000132063
|
si:dkey-1h24.6
|
si:dkey-1h24.6 |
| chr1_+_56972616 | 0.03 |
ENSDART00000152812
|
si:ch211-1f22.11
|
si:ch211-1f22.11 |
| chr4_-_52504961 | 0.03 |
ENSDART00000163123
|
si:dkey-78o7.3
|
si:dkey-78o7.3 |
| chr10_+_44955106 | 0.02 |
ENSDART00000185837
|
il1b
|
interleukin 1, beta |
| chr23_-_31645760 | 0.02 |
ENSDART00000035031
|
sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr4_-_53047883 | 0.02 |
ENSDART00000157496
|
si:dkey-201g16.5
|
si:dkey-201g16.5 |
| chr23_+_26102028 | 0.02 |
ENSDART00000139488
|
si:dkey-78k11.4
|
si:dkey-78k11.4 |
| chr1_+_57041549 | 0.02 |
ENSDART00000152198
|
si:ch211-1f22.16
|
si:ch211-1f22.16 |
| chr19_-_42416696 | 0.02 |
ENSDART00000086961
|
vps45
|
vacuolar protein sorting 45 homolog (S. cerevisiae) |
| chr4_+_35594200 | 0.02 |
ENSDART00000193902
|
si:dkeyp-4c4.2
|
si:dkeyp-4c4.2 |
| chr10_+_44986419 | 0.02 |
ENSDART00000162566
|
h2afvb
|
H2A histone family, member Vb |
| chr5_-_49011912 | 0.02 |
ENSDART00000097450
|
si:dkey-172m14.2
|
si:dkey-172m14.2 |
| chr4_+_31405646 | 0.02 |
ENSDART00000128732
|
si:rp71-5o12.3
|
si:rp71-5o12.3 |
| chr18_+_9641210 | 0.02 |
ENSDART00000182024
|
pclob
|
piccolo presynaptic cytomatrix protein b |
| chr14_+_18782610 | 0.02 |
ENSDART00000164468
|
si:ch211-111e20.1
|
si:ch211-111e20.1 |
| chr10_+_1899595 | 0.02 |
ENSDART00000040271
|
CU861476.1
|
|
| chr10_-_40968095 | 0.01 |
ENSDART00000184104
|
npffr1l1
|
neuropeptide FF receptor 1 like 1 |
| chr12_-_10441661 | 0.01 |
ENSDART00000014379
|
uqcrc2b
|
ubiquinol-cytochrome c reductase core protein 2b |
| chr14_-_33218418 | 0.01 |
ENSDART00000163046
|
si:dkey-31j3.11
|
si:dkey-31j3.11 |
| chr17_-_33414781 | 0.01 |
ENSDART00000142203
ENSDART00000034638 |
ccdc28a
|
coiled-coil domain containing 28A |
| chr2_+_42318012 | 0.01 |
ENSDART00000138137
|
ftr08
|
finTRIM family, member 8 |
| chr4_-_52379723 | 0.01 |
ENSDART00000168272
|
si:dkeyp-107f9.4
|
si:dkeyp-107f9.4 |
| chr9_+_7909490 | 0.01 |
ENSDART00000133063
ENSDART00000109288 |
myo16
|
myosin XVI |
| chr16_+_45746549 | 0.01 |
ENSDART00000190403
|
paqr6
|
progestin and adipoQ receptor family member VI |
| chr17_+_12658411 | 0.01 |
ENSDART00000139918
|
gpn1
|
GPN-loop GTPase 1 |
| chr16_-_14587332 | 0.01 |
ENSDART00000012479
|
dscc1
|
DNA replication and sister chromatid cohesion 1 |
| chr2_+_23790748 | 0.01 |
ENSDART00000041877
|
csrnp1a
|
cysteine-serine-rich nuclear protein 1a |
| chr3_+_22059066 | 0.01 |
ENSDART00000155739
|
kansl1b
|
KAT8 regulatory NSL complex subunit 1b |
| chr6_+_612330 | 0.01 |
ENSDART00000166872
ENSDART00000191758 |
kynu
|
kynureninase |
| chr2_-_24369087 | 0.01 |
ENSDART00000081237
|
plvapa
|
plasmalemma vesicle associated protein a |
| chr13_+_22104298 | 0.01 |
ENSDART00000115393
|
si:dkey-174i8.1
|
si:dkey-174i8.1 |
| chr3_-_40529782 | 0.00 |
ENSDART00000055194
ENSDART00000141737 |
actb2
|
actin, beta 2 |
| chr16_+_34046733 | 0.00 |
ENSDART00000058424
|
fam46ba
|
family with sequence similarity 46, member Ba |
Network of associatons between targets according to the STRING database.
First level regulatory network of nfat5a+nfat5b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 0.3 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 0.3 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.1 | 0.3 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.1 | 0.3 | GO:0043476 | pigment accumulation(GO:0043476) |
| 0.0 | 0.2 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 0.1 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.7 | GO:0006942 | regulation of striated muscle contraction(GO:0006942) |
| 0.0 | 0.1 | GO:0060547 | negative regulation of necroptotic process(GO:0060546) negative regulation of necrotic cell death(GO:0060547) |
| 0.0 | 0.2 | GO:0000305 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.1 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.2 | GO:0015800 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.4 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.3 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.4 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.2 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.3 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.1 | 0.7 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.1 | 0.4 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 1.4 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.5 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.2 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.2 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.1 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 0.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.1 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.0 | 0.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.2 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.0 | 0.2 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |