Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
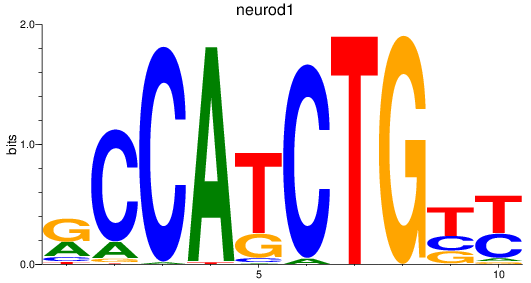
Results for neurod1
Z-value: 1.05
Transcription factors associated with neurod1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
neurod1
|
ENSDARG00000019566 | neuronal differentiation 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| neurod1 | dr11_v1_chr9_-_44295071_44295071 | 0.30 | 4.3e-01 | Click! |
Activity profile of neurod1 motif
Sorted Z-values of neurod1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_-_554142 | 0.84 |
ENSDART00000028997
|
myo9ab
|
myosin IXAb |
| chr3_-_50443607 | 0.61 |
ENSDART00000074036
|
rcvrna
|
recoverin a |
| chr11_-_28911172 | 0.57 |
ENSDART00000168493
|
igsf21a
|
immunoglobin superfamily, member 21a |
| chr9_-_1200187 | 0.56 |
ENSDART00000158760
|
ino80da
|
INO80 complex subunit Da |
| chr8_-_26609259 | 0.56 |
ENSDART00000027301
|
sema3b
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B |
| chr1_+_21937201 | 0.55 |
ENSDART00000087729
|
kdm4c
|
lysine (K)-specific demethylase 4C |
| chr12_+_21525496 | 0.53 |
ENSDART00000152974
|
ca10a
|
carbonic anhydrase Xa |
| chr2_+_50626476 | 0.52 |
ENSDART00000018150
|
neurod6b
|
neuronal differentiation 6b |
| chr21_-_13085242 | 0.49 |
ENSDART00000044504
|
zgc:109965
|
zgc:109965 |
| chr6_-_9565526 | 0.48 |
ENSDART00000151470
|
map3k2
|
mitogen-activated protein kinase kinase kinase 2 |
| chr9_+_25096500 | 0.47 |
ENSDART00000135074
ENSDART00000180436 ENSDART00000108629 |
lrch1
|
leucine-rich repeats and calponin homology (CH) domain containing 1 |
| chr5_+_62611400 | 0.46 |
ENSDART00000132054
|
abr
|
active BCR-related |
| chr7_+_29133321 | 0.46 |
ENSDART00000052346
|
gnao1b
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O, b |
| chr24_-_1657276 | 0.45 |
ENSDART00000168131
|
si:ch73-378g22.1
|
si:ch73-378g22.1 |
| chr6_-_13783604 | 0.41 |
ENSDART00000149536
ENSDART00000041269 ENSDART00000150102 |
cryba2a
|
crystallin, beta A2a |
| chr5_-_18513950 | 0.40 |
ENSDART00000145878
|
si:dkey-215k6.1
|
si:dkey-215k6.1 |
| chr2_+_31475772 | 0.38 |
ENSDART00000130722
|
cacnb2b
|
calcium channel, voltage-dependent, beta 2b |
| chr9_-_44295071 | 0.37 |
ENSDART00000011837
|
neurod1
|
neuronal differentiation 1 |
| chr2_+_7557912 | 0.37 |
ENSDART00000160053
|
ripk2
|
receptor-interacting serine-threonine kinase 2 |
| chr5_-_19932621 | 0.36 |
ENSDART00000088881
|
git2a
|
G protein-coupled receptor kinase interacting ArfGAP 2a |
| chr16_-_33097398 | 0.36 |
ENSDART00000166617
|
dopey1
|
dopey family member 1 |
| chr19_-_38872650 | 0.36 |
ENSDART00000146641
|
adgrb2
|
adhesion G protein-coupled receptor B2 |
| chr18_+_46773198 | 0.35 |
ENSDART00000174647
|
CABZ01041495.1
|
|
| chr16_-_29334672 | 0.35 |
ENSDART00000162835
|
bcan
|
brevican |
| chr17_-_25397558 | 0.35 |
ENSDART00000182052
|
fam167b
|
family with sequence similarity 167, member B |
| chr18_+_45550783 | 0.34 |
ENSDART00000138075
|
kifc3
|
kinesin family member C3 |
| chr23_+_20563779 | 0.34 |
ENSDART00000146008
|
camkvl
|
CaM kinase-like vesicle-associated, like |
| chr3_-_21061931 | 0.33 |
ENSDART00000036741
|
fam57ba
|
family with sequence similarity 57, member Ba |
| chr3_-_21062706 | 0.33 |
ENSDART00000155605
ENSDART00000153686 ENSDART00000157168 ENSDART00000156614 ENSDART00000155743 ENSDART00000156275 |
fam57ba
|
family with sequence similarity 57, member Ba |
| chr5_+_11407504 | 0.32 |
ENSDART00000186723
ENSDART00000018047 |
zgc:112294
|
zgc:112294 |
| chr3_-_60886984 | 0.32 |
ENSDART00000170974
|
PITPNC1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr7_-_48263516 | 0.32 |
ENSDART00000006619
ENSDART00000142370 ENSDART00000148273 ENSDART00000147968 |
rbpms2b
|
RNA binding protein with multiple splicing 2b |
| chr12_-_14922955 | 0.31 |
ENSDART00000002078
|
neurod2
|
neurogenic differentiation 2 |
| chr9_-_22831836 | 0.31 |
ENSDART00000142585
|
neb
|
nebulin |
| chr23_+_5470967 | 0.31 |
ENSDART00000110522
ENSDART00000149050 |
tulp1a
|
tubby like protein 1a |
| chr11_+_30647545 | 0.31 |
ENSDART00000114792
|
gb:eh507706
|
expressed sequence EH507706 |
| chr3_+_50069610 | 0.30 |
ENSDART00000056619
|
zgc:103625
|
zgc:103625 |
| chr12_-_22238004 | 0.30 |
ENSDART00000038310
|
ormdl3
|
ORMDL sphingolipid biosynthesis regulator 3 |
| chr12_+_23892972 | 0.30 |
ENSDART00000152852
|
svila
|
supervillin a |
| chr1_-_34450784 | 0.30 |
ENSDART00000140515
|
lmo7b
|
LIM domain 7b |
| chr12_-_41618844 | 0.30 |
ENSDART00000160054
|
dpysl4
|
dihydropyrimidinase-like 4 |
| chr8_+_36500061 | 0.29 |
ENSDART00000185840
|
slc7a4
|
solute carrier family 7, member 4 |
| chr6_+_18359306 | 0.29 |
ENSDART00000157796
|
cbx8b
|
chromobox homolog 8b |
| chr6_-_26225814 | 0.29 |
ENSDART00000089121
|
hs2st1b
|
heparan sulfate 2-O-sulfotransferase 1b |
| chr5_-_33769211 | 0.29 |
ENSDART00000133504
|
dab2ipb
|
DAB2 interacting protein b |
| chr13_-_7031033 | 0.29 |
ENSDART00000193211
|
CABZ01061524.1
|
|
| chr5_+_36768674 | 0.29 |
ENSDART00000146854
|
mark4a
|
MAP/microtubule affinity-regulating kinase 4a |
| chr19_-_10656667 | 0.29 |
ENSDART00000081379
ENSDART00000151456 ENSDART00000143271 ENSDART00000182126 |
olah
|
oleoyl-ACP hydrolase |
| chr16_-_29480335 | 0.28 |
ENSDART00000148930
|
lingo4b
|
leucine rich repeat and Ig domain containing 4b |
| chr21_+_6212844 | 0.28 |
ENSDART00000150301
|
fnbp1b
|
formin binding protein 1b |
| chr7_+_50828774 | 0.28 |
ENSDART00000182821
|
per1b
|
period circadian clock 1b |
| chr24_+_40905100 | 0.28 |
ENSDART00000167854
|
scn12ab
|
sodium channel, voltage gated, type XII, alpha b |
| chr19_-_38830582 | 0.27 |
ENSDART00000189966
ENSDART00000183055 |
adgrb2
|
adhesion G protein-coupled receptor B2 |
| chr6_-_9563534 | 0.27 |
ENSDART00000184765
|
map3k2
|
mitogen-activated protein kinase kinase kinase 2 |
| chr6_+_13787855 | 0.27 |
ENSDART00000182899
|
tmem198b
|
transmembrane protein 198b |
| chr1_-_40227166 | 0.27 |
ENSDART00000146680
|
si:ch211-113e8.3
|
si:ch211-113e8.3 |
| chr19_-_32888758 | 0.27 |
ENSDART00000052080
|
laptm4b
|
lysosomal protein transmembrane 4 beta |
| chr12_-_41619257 | 0.27 |
ENSDART00000162967
|
dpysl4
|
dihydropyrimidinase-like 4 |
| chr7_+_30823749 | 0.27 |
ENSDART00000085661
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr20_+_23498255 | 0.27 |
ENSDART00000149922
|
palld
|
palladin, cytoskeletal associated protein |
| chr2_-_32574944 | 0.27 |
ENSDART00000056642
|
tmub1
|
transmembrane and ubiquitin-like domain containing 1 |
| chr12_+_46543572 | 0.26 |
ENSDART00000167510
|
hid1b
|
HID1 domain containing b |
| chr15_+_30158652 | 0.26 |
ENSDART00000190682
|
nlk2
|
nemo-like kinase, type 2 |
| chr23_-_18057851 | 0.26 |
ENSDART00000173075
ENSDART00000173230 ENSDART00000173135 ENSDART00000173431 ENSDART00000173068 ENSDART00000172987 |
zgc:92287
|
zgc:92287 |
| chr5_-_40190949 | 0.26 |
ENSDART00000175588
|
wdfy3
|
WD repeat and FYVE domain containing 3 |
| chr5_+_60919378 | 0.26 |
ENSDART00000184915
|
doc2b
|
double C2-like domains, beta |
| chr23_+_5465806 | 0.26 |
ENSDART00000149434
ENSDART00000148506 |
tulp1a
|
tubby like protein 1a |
| chr24_-_10828560 | 0.25 |
ENSDART00000132282
|
fam49bb
|
family with sequence similarity 49, member Bb |
| chr17_+_12075805 | 0.25 |
ENSDART00000155329
|
cnsta
|
consortin, connexin sorting protein a |
| chr23_-_18057553 | 0.25 |
ENSDART00000173102
ENSDART00000058742 |
zgc:92287
|
zgc:92287 |
| chr2_-_22230326 | 0.25 |
ENSDART00000127810
|
fam110b
|
family with sequence similarity 110, member B |
| chr24_-_17444067 | 0.25 |
ENSDART00000155843
|
cntnap2a
|
contactin associated protein like 2a |
| chr21_+_39100289 | 0.25 |
ENSDART00000075958
|
slc13a2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr14_+_32838110 | 0.24 |
ENSDART00000158077
|
arr3b
|
arrestin 3b, retinal (X-arrestin) |
| chr19_-_5812319 | 0.24 |
ENSDART00000114472
|
si:ch211-264f5.8
|
si:ch211-264f5.8 |
| chr20_-_44576949 | 0.24 |
ENSDART00000148639
|
ubxn2a
|
UBX domain protein 2A |
| chr20_-_18731268 | 0.24 |
ENSDART00000183893
|
enpp5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 |
| chr21_-_16632808 | 0.24 |
ENSDART00000172645
|
unc5da
|
unc-5 netrin receptor Da |
| chr19_-_6134802 | 0.24 |
ENSDART00000140051
|
cica
|
capicua transcriptional repressor a |
| chr8_+_7359294 | 0.24 |
ENSDART00000121708
|
pcsk1nl
|
proprotein convertase subtilisin/kexin type 1 inhibitor, like |
| chr12_+_31744217 | 0.24 |
ENSDART00000190361
|
RNF157
|
si:dkey-49c17.3 |
| chr7_+_39389273 | 0.24 |
ENSDART00000191298
|
tnni2b.2
|
troponin I type 2b (skeletal, fast), tandem duplicate 2 |
| chr7_+_55950229 | 0.23 |
ENSDART00000082780
|
ACSF3
|
acyl-CoA synthetase family member 3 |
| chr24_+_32472155 | 0.23 |
ENSDART00000098859
|
neurod6a
|
neuronal differentiation 6a |
| chr2_+_5948534 | 0.23 |
ENSDART00000124324
ENSDART00000176461 |
slc1a7a
|
solute carrier family 1 (glutamate transporter), member 7a |
| chr10_+_15777258 | 0.23 |
ENSDART00000140511
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr18_+_29145681 | 0.23 |
ENSDART00000089031
ENSDART00000193336 |
ppfibp2a
|
PTPRF interacting protein, binding protein 2a (liprin beta 2) |
| chr6_-_35487413 | 0.23 |
ENSDART00000102461
|
rgs8
|
regulator of G protein signaling 8 |
| chr18_-_12858016 | 0.23 |
ENSDART00000130343
|
parp12a
|
poly (ADP-ribose) polymerase family, member 12a |
| chr8_-_6943155 | 0.23 |
ENSDART00000139545
ENSDART00000033294 |
wdr13
|
WD repeat domain 13 |
| chr14_+_32837914 | 0.23 |
ENSDART00000158888
|
arr3b
|
arrestin 3b, retinal (X-arrestin) |
| chr5_+_37854685 | 0.23 |
ENSDART00000051222
ENSDART00000185283 |
ins
|
preproinsulin |
| chr2_+_25198648 | 0.23 |
ENSDART00000110922
|
ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr5_-_26493253 | 0.23 |
ENSDART00000088222
|
arvcfb
|
ARVCF, delta catenin family member b |
| chr13_-_16257848 | 0.23 |
ENSDART00000079745
|
zgc:110045
|
zgc:110045 |
| chr13_-_16226312 | 0.22 |
ENSDART00000163952
|
zgc:110045
|
zgc:110045 |
| chr17_-_51829310 | 0.22 |
ENSDART00000154544
|
numb
|
numb homolog (Drosophila) |
| chr21_+_44300689 | 0.22 |
ENSDART00000186298
ENSDART00000142810 |
gabra3
|
gamma-aminobutyric acid (GABA) A receptor, alpha 3 |
| chr5_-_30145939 | 0.22 |
ENSDART00000086795
|
zbtb44
|
zinc finger and BTB domain containing 44 |
| chr5_+_42912966 | 0.22 |
ENSDART00000039973
|
rufy3
|
RUN and FYVE domain containing 3 |
| chr2_+_21309272 | 0.22 |
ENSDART00000141322
|
zbtb47a
|
zinc finger and BTB domain containing 47a |
| chr20_-_53963515 | 0.22 |
ENSDART00000110252
|
ppp2r5cb
|
protein phosphatase 2, regulatory subunit B', gamma b |
| chr24_-_34335265 | 0.22 |
ENSDART00000128690
|
agap3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr14_+_21722235 | 0.22 |
ENSDART00000183667
|
stx3a
|
syntaxin 3A |
| chr18_-_10995410 | 0.22 |
ENSDART00000136751
|
tspan33b
|
tetraspanin 33b |
| chr6_-_43449013 | 0.22 |
ENSDART00000122423
|
eevs
|
2-epi-5-epi-valiolone synthase |
| chr19_+_177455 | 0.22 |
ENSDART00000111580
|
tmem65
|
transmembrane protein 65 |
| chr16_-_51151993 | 0.22 |
ENSDART00000156255
|
ago1
|
argonaute RISC catalytic component 1 |
| chr13_-_44285793 | 0.22 |
ENSDART00000167383
|
CABZ01069436.1
|
|
| chr19_-_25427255 | 0.21 |
ENSDART00000036854
|
glcci1a
|
glucocorticoid induced 1a |
| chr12_-_35949936 | 0.21 |
ENSDART00000192583
|
AL954682.1
|
|
| chr19_+_43684376 | 0.21 |
ENSDART00000051723
|
si:ch211-193k19.1
|
si:ch211-193k19.1 |
| chr15_-_28094256 | 0.21 |
ENSDART00000142041
ENSDART00000132153 ENSDART00000146657 ENSDART00000048720 |
cryba1a
|
crystallin, beta A1a |
| chr11_+_24925434 | 0.21 |
ENSDART00000131431
|
sulf2a
|
sulfatase 2a |
| chr16_-_35329803 | 0.21 |
ENSDART00000161729
ENSDART00000157700 ENSDART00000184584 ENSDART00000174713 ENSDART00000162518 |
ptprub
|
protein tyrosine phosphatase, receptor type, U, b |
| chr6_-_4214297 | 0.21 |
ENSDART00000191433
|
trak2
|
trafficking protein, kinesin binding 2 |
| chr20_-_39273987 | 0.20 |
ENSDART00000127173
|
clu
|
clusterin |
| chr7_-_55292116 | 0.20 |
ENSDART00000122603
|
rnf166
|
ring finger protein 166 |
| chr13_-_30027730 | 0.20 |
ENSDART00000044009
|
scdb
|
stearoyl-CoA desaturase b |
| chr5_-_40734045 | 0.20 |
ENSDART00000010896
|
isl1
|
ISL LIM homeobox 1 |
| chr7_-_54217547 | 0.20 |
ENSDART00000162777
ENSDART00000188268 ENSDART00000165875 |
csnk1g1
|
casein kinase 1, gamma 1 |
| chr1_+_18863060 | 0.20 |
ENSDART00000139241
|
rnf38
|
ring finger protein 38 |
| chr17_-_36988455 | 0.20 |
ENSDART00000187180
ENSDART00000126823 |
dnmt3ab
|
DNA (cytosine-5-)-methyltransferase 3 alpha b |
| chr8_-_16592491 | 0.20 |
ENSDART00000101655
|
calr
|
calreticulin |
| chr6_-_51101834 | 0.20 |
ENSDART00000092493
|
ptprt
|
protein tyrosine phosphatase, receptor type, t |
| chr25_+_33849647 | 0.20 |
ENSDART00000121449
|
roraa
|
RAR-related orphan receptor A, paralog a |
| chr19_+_49721 | 0.20 |
ENSDART00000160489
|
col14a1b
|
collagen, type XIV, alpha 1b |
| chr25_-_35664817 | 0.20 |
ENSDART00000148718
|
lrrk2
|
leucine-rich repeat kinase 2 |
| chr22_+_24157807 | 0.20 |
ENSDART00000159165
|
b3galt2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
| chr1_-_29045426 | 0.20 |
ENSDART00000019770
|
gpm6ba
|
glycoprotein M6Ba |
| chr6_-_35032792 | 0.20 |
ENSDART00000168256
|
ddr2a
|
discoidin domain receptor tyrosine kinase 2a |
| chr12_-_33659328 | 0.20 |
ENSDART00000153457
|
tmem94
|
transmembrane protein 94 |
| chr6_+_52804267 | 0.20 |
ENSDART00000065681
|
matn4
|
matrilin 4 |
| chr24_+_14801844 | 0.19 |
ENSDART00000141620
|
pi15a
|
peptidase inhibitor 15a |
| chr21_+_8427059 | 0.19 |
ENSDART00000143151
|
dennd1a
|
DENN/MADD domain containing 1A |
| chr2_-_21352101 | 0.19 |
ENSDART00000057021
|
hhatla
|
hedgehog acyltransferase like, a |
| chr18_-_42313798 | 0.19 |
ENSDART00000098639
|
cntn5
|
contactin 5 |
| chr12_+_32292564 | 0.19 |
ENSDART00000152945
|
ANKFN1
|
si:ch211-277e21.2 |
| chr10_-_5847904 | 0.19 |
ENSDART00000161096
|
ankrd55
|
ankyrin repeat domain 55 |
| chr19_+_33553586 | 0.19 |
ENSDART00000183477
|
runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr10_+_15777064 | 0.19 |
ENSDART00000114483
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr17_+_23146976 | 0.19 |
ENSDART00000114212
|
rasgrp3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr25_-_18948816 | 0.19 |
ENSDART00000091549
|
nt5dc3
|
5'-nucleotidase domain containing 3 |
| chr19_-_868187 | 0.18 |
ENSDART00000186626
|
eomesa
|
eomesodermin homolog a |
| chr23_-_31647793 | 0.18 |
ENSDART00000145621
|
sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr3_-_61494840 | 0.18 |
ENSDART00000101957
|
baiap2l1b
|
BAI1-associated protein 2-like 1b |
| chr6_+_22597362 | 0.18 |
ENSDART00000131242
|
cygb2
|
cytoglobin 2 |
| chr20_+_6756247 | 0.18 |
ENSDART00000167344
|
igfbp3
|
insulin-like growth factor binding protein 3 |
| chr23_-_18057270 | 0.18 |
ENSDART00000173385
|
zgc:92287
|
zgc:92287 |
| chr6_+_9175886 | 0.18 |
ENSDART00000165333
|
si:ch211-207l14.1
|
si:ch211-207l14.1 |
| chr10_+_31809226 | 0.18 |
ENSDART00000087898
|
foxo1b
|
forkhead box O1 b |
| chr8_-_50147948 | 0.18 |
ENSDART00000149010
|
hp
|
haptoglobin |
| chr20_+_48100261 | 0.18 |
ENSDART00000158604
|
xkr5a
|
XK related 5a |
| chr19_-_24136233 | 0.18 |
ENSDART00000143365
|
thap7
|
THAP domain containing 7 |
| chr18_-_399554 | 0.18 |
ENSDART00000164374
ENSDART00000186311 ENSDART00000181816 ENSDART00000181892 |
si:ch211-79l17.1
|
si:ch211-79l17.1 |
| chr1_-_14233815 | 0.18 |
ENSDART00000044896
|
camk2d2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 2 |
| chr3_-_36745935 | 0.18 |
ENSDART00000184853
|
abcc6b.1
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 6b, tandem duplicate 1 |
| chr17_+_25397070 | 0.18 |
ENSDART00000164254
|
zgc:154055
|
zgc:154055 |
| chr7_+_74134010 | 0.18 |
ENSDART00000164874
|
cldnd1a
|
claudin domain containing 1a |
| chr10_-_6867282 | 0.17 |
ENSDART00000144001
ENSDART00000109744 |
ppip5k2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr8_+_25145464 | 0.17 |
ENSDART00000136505
|
gnai3
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3 |
| chr10_+_29431529 | 0.17 |
ENSDART00000158154
|
dlg2
|
discs, large homolog 2 (Drosophila) |
| chr17_-_2535682 | 0.17 |
ENSDART00000155227
|
ccdc9b
|
coiled-coil domain containing 9B |
| chr17_-_51202339 | 0.17 |
ENSDART00000167117
|
si:ch1073-469d17.2
|
si:ch1073-469d17.2 |
| chr13_+_8604710 | 0.17 |
ENSDART00000091097
|
socs5b
|
suppressor of cytokine signaling 5b |
| chr20_+_21268795 | 0.17 |
ENSDART00000090016
|
nudt14
|
nudix (nucleoside diphosphate linked moiety X)-type motif 14 |
| chr18_-_38087875 | 0.17 |
ENSDART00000111301
|
luzp2
|
leucine zipper protein 2 |
| chr8_-_49935358 | 0.17 |
ENSDART00000159782
ENSDART00000156841 |
agtpbp1
CU694202.1
|
ATP/GTP binding protein 1 |
| chr22_-_33679277 | 0.17 |
ENSDART00000169948
|
FO904977.1
|
|
| chr4_-_1324141 | 0.17 |
ENSDART00000180720
|
ptn
|
pleiotrophin |
| chr7_+_26224211 | 0.17 |
ENSDART00000173999
|
vgf
|
VGF nerve growth factor inducible |
| chr15_-_20954552 | 0.17 |
ENSDART00000006910
|
tbcela
|
tubulin folding cofactor E-like a |
| chr5_-_51819027 | 0.17 |
ENSDART00000164267
|
homer1b
|
homer scaffolding protein 1b |
| chr9_+_49712868 | 0.17 |
ENSDART00000192969
ENSDART00000183310 |
CSRNP3
|
cysteine and serine rich nuclear protein 3 |
| chr21_-_43527198 | 0.17 |
ENSDART00000126092
|
irs4a
|
insulin receptor substrate 4a |
| chr1_-_7893808 | 0.17 |
ENSDART00000110154
|
radil
|
Ras association and DIL domains |
| chr18_+_15795566 | 0.17 |
ENSDART00000099944
|
nudt4a
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4a |
| chr23_-_27692717 | 0.17 |
ENSDART00000053878
ENSDART00000145028 |
IKZF4
|
si:dkey-166n8.9 |
| chr3_+_36972298 | 0.17 |
ENSDART00000150917
|
si:ch211-18i17.2
|
si:ch211-18i17.2 |
| chr23_+_25172682 | 0.17 |
ENSDART00000191197
ENSDART00000183497 |
si:dkey-151g10.3
|
si:dkey-151g10.3 |
| chr11_+_6881001 | 0.16 |
ENSDART00000170331
|
klhl26
|
kelch-like family member 26 |
| chr7_+_32722227 | 0.16 |
ENSDART00000126565
|
si:ch211-150g13.3
|
si:ch211-150g13.3 |
| chr13_-_12667220 | 0.16 |
ENSDART00000079594
|
fam241a
|
family with sequence similarity 241 member A |
| chr15_-_7598294 | 0.16 |
ENSDART00000165898
|
gbe1b
|
glucan (1,4-alpha-), branching enzyme 1b |
| chr20_-_26001288 | 0.16 |
ENSDART00000136518
ENSDART00000063177 |
capn3b
|
calpain 3b |
| chr4_+_8376362 | 0.16 |
ENSDART00000138653
ENSDART00000132647 |
erc1b
|
ELKS/RAB6-interacting/CAST family member 1b |
| chr6_+_21095918 | 0.16 |
ENSDART00000167225
|
spega
|
SPEG complex locus a |
| chr7_-_58130703 | 0.16 |
ENSDART00000172082
|
ank2b
|
ankyrin 2b, neuronal |
| chr11_+_6902946 | 0.16 |
ENSDART00000144006
|
crtc1b
|
CREB regulated transcription coactivator 1b |
| chr11_+_37049347 | 0.16 |
ENSDART00000109235
|
bicd2
|
bicaudal D homolog 2 (Drosophila) |
| chr5_-_38342992 | 0.16 |
ENSDART00000140337
|
mink1
|
misshapen-like kinase 1 |
| chr21_+_27382893 | 0.16 |
ENSDART00000005682
|
actn3a
|
actinin alpha 3a |
| chr21_+_13353263 | 0.16 |
ENSDART00000114677
|
si:ch73-62l21.1
|
si:ch73-62l21.1 |
| chr2_-_31800521 | 0.16 |
ENSDART00000112763
|
retreg1
|
reticulophagy regulator 1 |
| chr11_+_37049105 | 0.16 |
ENSDART00000155408
|
bicd2
|
bicaudal D homolog 2 (Drosophila) |
| chr20_+_1412193 | 0.16 |
ENSDART00000064419
|
leg1.1
|
liver-enriched gene 1, tandem duplicate 1 |
| chr16_-_32013913 | 0.16 |
ENSDART00000030282
ENSDART00000138701 |
gstk1
|
glutathione S-transferase kappa 1 |
| chr3_+_36972586 | 0.16 |
ENSDART00000102784
|
si:ch211-18i17.2
|
si:ch211-18i17.2 |
| chr21_-_22737228 | 0.16 |
ENSDART00000151366
|
fbxo40.2
|
F-box protein 40, tandem duplicate 2 |
| chr8_-_3716734 | 0.16 |
ENSDART00000172966
|
bicdl1
|
BICD family like cargo adaptor 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of neurod1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0098529 | neuromuscular junction development, skeletal muscle fiber(GO:0098529) |
| 0.2 | 0.5 | GO:1990868 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) negative regulation of lymphocyte migration(GO:2000402) negative regulation of T cell migration(GO:2000405) |
| 0.1 | 0.4 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.1 | 0.3 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.1 | 0.3 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.1 | 0.3 | GO:0090155 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.1 | 0.2 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.1 | 0.4 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.1 | 0.2 | GO:0051580 | regulation of neurotransmitter uptake(GO:0051580) negative regulation of anion transport(GO:1903792) |
| 0.1 | 0.2 | GO:0060262 | N-terminal protein palmitoylation(GO:0006500) negative regulation of lipoprotein metabolic process(GO:0050748) regulation of N-terminal protein palmitoylation(GO:0060254) negative regulation of N-terminal protein palmitoylation(GO:0060262) negative regulation of protein lipidation(GO:1903060) |
| 0.1 | 0.3 | GO:0051145 | smooth muscle cell differentiation(GO:0051145) negative regulation of muscle cell differentiation(GO:0051148) |
| 0.1 | 0.3 | GO:1901911 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 0.2 | GO:0060571 | invagination involved in gastrulation with mouth forming second(GO:0055109) morphogenesis of an epithelial fold(GO:0060571) |
| 0.1 | 0.2 | GO:0030858 | positive regulation of epithelial cell differentiation(GO:0030858) negative regulation of behavior(GO:0048521) negative regulation of feeding behavior(GO:2000252) |
| 0.1 | 0.6 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.1 | 0.2 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 0.4 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.1 | 0.2 | GO:0006145 | allantoin catabolic process(GO:0000256) purine nucleobase catabolic process(GO:0006145) |
| 0.1 | 0.3 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.1 | 0.2 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.1 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.0 | 0.2 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.0 | 0.2 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.0 | 0.2 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.5 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.0 | 0.1 | GO:0036076 | ligamentous ossification(GO:0036076) |
| 0.0 | 0.2 | GO:0055107 | Golgi to secretory granule transport(GO:0055107) |
| 0.0 | 0.3 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.1 | GO:0010898 | regulation of triglyceride catabolic process(GO:0010896) positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.0 | 0.1 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.1 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.2 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.3 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.3 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.0 | 0.6 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) nucleobase catabolic process(GO:0046113) |
| 0.0 | 0.1 | GO:0006589 | octopamine biosynthetic process(GO:0006589) dopamine catabolic process(GO:0042420) norepinephrine biosynthetic process(GO:0042421) octopamine metabolic process(GO:0046333) |
| 0.0 | 0.4 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.0 | 0.2 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.2 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.3 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 0.1 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.1 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.0 | 0.1 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.0 | 0.3 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.3 | GO:0048790 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.0 | 0.1 | GO:0003091 | renal water homeostasis(GO:0003091) renal water transport(GO:0003097) |
| 0.0 | 0.2 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.5 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.1 | GO:0032447 | tRNA wobble position uridine thiolation(GO:0002143) protein urmylation(GO:0032447) |
| 0.0 | 0.2 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.3 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.1 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.0 | 0.1 | GO:0044241 | lipid digestion(GO:0044241) |
| 0.0 | 0.2 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.1 | GO:0006921 | apoptotic DNA fragmentation(GO:0006309) cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.0 | 0.5 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.2 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.1 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.3 | GO:0048923 | posterior lateral line neuromast hair cell differentiation(GO:0048923) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.6 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.0 | 0.4 | GO:0007257 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 0.1 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.2 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.4 | GO:0051928 | positive regulation of calcium ion transport(GO:0051928) |
| 0.0 | 0.2 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.0 | 0.2 | GO:1990402 | embryonic liver development(GO:1990402) |
| 0.0 | 0.1 | GO:2000479 | regulation of cAMP-dependent protein kinase activity(GO:2000479) |
| 0.0 | 0.2 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0055062 | phosphate ion homeostasis(GO:0055062) trivalent inorganic anion homeostasis(GO:0072506) |
| 0.0 | 0.7 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 0.2 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.2 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.2 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.0 | GO:0034773 | histone H4-K20 methylation(GO:0034770) histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.0 | GO:0019284 | L-methionine biosynthetic process from S-adenosylmethionine(GO:0019284) |
| 0.0 | 0.1 | GO:0055117 | regulation of cardiac muscle contraction(GO:0055117) |
| 0.0 | 0.1 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.0 | 0.1 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.0 | 0.5 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.5 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.1 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.0 | 0.2 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 0.1 | GO:1904292 | regulation of ERAD pathway(GO:1904292) |
| 0.0 | 0.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.0 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.1 | GO:0043201 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.0 | 0.4 | GO:1903039 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.0 | 0.2 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 0.7 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.2 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 0.3 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.1 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 0.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.2 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.3 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.6 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.8 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.1 | GO:0038039 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.1 | GO:0008247 | 1-alkyl-2-acetylglycerophosphocholine esterase complex(GO:0008247) |
| 0.0 | 0.3 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.3 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.3 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) |
| 0.0 | 0.0 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.0 | 0.1 | GO:0061700 | GATOR2 complex(GO:0061700) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004394 | heparan sulfate 2-O-sulfotransferase activity(GO:0004394) |
| 0.1 | 0.5 | GO:0051430 | mu-type opioid receptor binding(GO:0031852) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 0.3 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 0.3 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.5 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 0.2 | GO:0032896 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.0 | 0.6 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.2 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.2 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.4 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.2 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 0.0 | 0.6 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.2 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.6 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.1 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) pre-miRNA binding(GO:0070883) |
| 0.0 | 0.1 | GO:0016436 | rRNA (uridine) methyltransferase activity(GO:0016436) |
| 0.0 | 0.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.3 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.2 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.1 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.0 | 0.3 | GO:0031995 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 0.7 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.2 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.0 | 0.1 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.2 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.0 | 0.2 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.2 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) |
| 0.0 | 0.1 | GO:0005009 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.1 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.0 | 0.2 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.2 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.2 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.1 | GO:0004775 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.0 | 0.1 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.0 | 0.1 | GO:0031841 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.0 | 0.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.6 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.1 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.3 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0009384 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) N-acylmannosamine kinase activity(GO:0009384) |
| 0.0 | 0.2 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.8 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.2 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.1 | GO:0008887 | glycerate kinase activity(GO:0008887) |
| 0.0 | 0.1 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.2 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.0 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.2 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.2 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.2 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.2 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.1 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.2 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.1 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.3 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.0 | 0.3 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.3 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.6 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.5 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.2 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.2 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.4 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.1 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.4 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.4 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.1 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.1 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.1 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.2 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |