Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
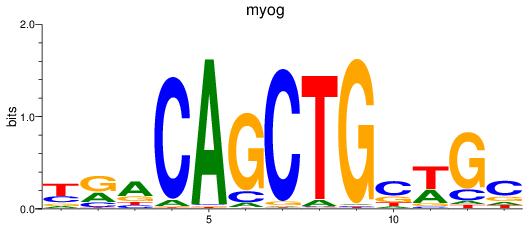
Results for myog
Z-value: 1.16
Transcription factors associated with myog
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
myog
|
ENSDARG00000009438 | myogenin |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| myog | dr11_v1_chr11_-_22599584_22599584 | 0.78 | 1.2e-02 | Click! |
Activity profile of myog motif
Sorted Z-values of myog motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_61203203 | 2.45 |
ENSDART00000171787
|
pvalb1
|
parvalbumin 1 |
| chr3_+_26145013 | 2.00 |
ENSDART00000162546
ENSDART00000129561 |
atp2a1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr7_+_44715224 | 1.90 |
ENSDART00000184630
|
si:dkey-56m19.5
|
si:dkey-56m19.5 |
| chr11_-_5865744 | 1.88 |
ENSDART00000104360
|
gamt
|
guanidinoacetate N-methyltransferase |
| chr21_+_27382893 | 1.87 |
ENSDART00000005682
|
actn3a
|
actinin alpha 3a |
| chr7_+_29951997 | 1.67 |
ENSDART00000173453
|
tpma
|
alpha-tropomyosin |
| chr5_-_19400166 | 1.56 |
ENSDART00000008994
|
foxn4
|
forkhead box N4 |
| chr22_-_57177 | 1.46 |
ENSDART00000163959
|
CABZ01085139.1
|
|
| chr3_+_26144765 | 1.42 |
ENSDART00000146267
ENSDART00000043932 |
atp2a1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr16_+_23947196 | 1.42 |
ENSDART00000103190
ENSDART00000132961 ENSDART00000147690 ENSDART00000142168 |
apoa4b.2
|
apolipoprotein A-IV b, tandem duplicate 2 |
| chr7_+_49715750 | 1.39 |
ENSDART00000019446
|
ascl1b
|
achaete-scute family bHLH transcription factor 1b |
| chr6_-_39313027 | 1.37 |
ENSDART00000012644
|
krt4
|
keratin 4 |
| chr11_-_41966854 | 1.35 |
ENSDART00000055709
|
her2
|
hairy-related 2 |
| chr6_-_42003780 | 1.35 |
ENSDART00000032527
|
cav3
|
caveolin 3 |
| chr11_-_55224 | 1.32 |
ENSDART00000159169
|
col2a1b
|
collagen, type II, alpha 1b |
| chr2_+_42191592 | 1.31 |
ENSDART00000144716
|
cavin4a
|
caveolae associated protein 4a |
| chr5_-_64203101 | 1.23 |
ENSDART00000029364
|
ak5l
|
adenylate kinase 5, like |
| chr16_+_23921777 | 1.23 |
ENSDART00000163213
|
apoa4b.3
|
apolipoprotein A-IV b, tandem duplicate 3 |
| chr16_+_23921610 | 1.22 |
ENSDART00000143855
|
apoa4b.3
|
apolipoprotein A-IV b, tandem duplicate 3 |
| chr2_-_7666021 | 1.19 |
ENSDART00000180007
|
CABZ01021592.1
|
|
| chr7_+_39444843 | 1.19 |
ENSDART00000143999
ENSDART00000173554 ENSDART00000173698 ENSDART00000173754 ENSDART00000144075 ENSDART00000138192 ENSDART00000145457 ENSDART00000141750 ENSDART00000103056 ENSDART00000142946 ENSDART00000173748 |
tnnt3b
|
troponin T type 3b (skeletal, fast) |
| chr21_+_5129513 | 1.18 |
ENSDART00000102572
|
thbs4b
|
thrombospondin 4b |
| chr11_+_7158723 | 1.17 |
ENSDART00000035560
|
tmem38a
|
transmembrane protein 38A |
| chr22_+_20720808 | 1.16 |
ENSDART00000171321
|
si:dkey-211f22.5
|
si:dkey-211f22.5 |
| chr7_-_74090168 | 1.13 |
ENSDART00000050528
|
tyrp1a
|
tyrosinase-related protein 1a |
| chr2_-_21335131 | 1.10 |
ENSDART00000057022
|
klhl40a
|
kelch-like family member 40a |
| chr4_-_16354292 | 1.09 |
ENSDART00000139919
|
lum
|
lumican |
| chr3_-_50865079 | 1.08 |
ENSDART00000164295
|
pmp22a
|
peripheral myelin protein 22a |
| chr11_-_101758 | 1.08 |
ENSDART00000173015
|
elmo2
|
engulfment and cell motility 2 |
| chr17_+_52822831 | 1.06 |
ENSDART00000193368
|
meis2a
|
Meis homeobox 2a |
| chr20_-_44496245 | 1.04 |
ENSDART00000012229
|
fkbp1b
|
FK506 binding protein 1b |
| chr10_+_9550419 | 1.01 |
ENSDART00000064977
|
si:ch211-243g18.2
|
si:ch211-243g18.2 |
| chr4_-_4834347 | 1.00 |
ENSDART00000141803
|
coa6
|
cytochrome c oxidase assembly factor 6 |
| chr24_+_20575259 | 0.99 |
ENSDART00000010488
|
klhl40b
|
kelch-like family member 40b |
| chr6_-_32703317 | 0.97 |
ENSDART00000064833
|
mafaa
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Aa |
| chr8_+_44926946 | 0.95 |
ENSDART00000098567
|
zgc:154046
|
zgc:154046 |
| chr11_-_21030070 | 0.94 |
ENSDART00000186322
|
fmoda
|
fibromodulin a |
| chr19_-_5254699 | 0.94 |
ENSDART00000081951
|
stx1b
|
syntaxin 1B |
| chr6_+_40629066 | 0.93 |
ENSDART00000103757
|
slc6a11a
|
solute carrier family 6 (neurotransmitter transporter), member 11a |
| chr5_+_37854685 | 0.93 |
ENSDART00000051222
ENSDART00000185283 |
ins
|
preproinsulin |
| chr4_-_5597802 | 0.92 |
ENSDART00000136229
|
vegfab
|
vascular endothelial growth factor Ab |
| chr23_-_45405968 | 0.92 |
ENSDART00000149462
|
zgc:101853
|
zgc:101853 |
| chr23_+_22656477 | 0.91 |
ENSDART00000009337
ENSDART00000133322 |
eno1a
|
enolase 1a, (alpha) |
| chr13_-_31622195 | 0.90 |
ENSDART00000057432
|
six1a
|
SIX homeobox 1a |
| chr20_+_35382482 | 0.89 |
ENSDART00000135284
|
vsnl1a
|
visinin-like 1a |
| chr4_-_16353733 | 0.88 |
ENSDART00000186785
|
lum
|
lumican |
| chr23_-_3409140 | 0.87 |
ENSDART00000002309
|
mafba
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Ba |
| chr1_-_20271138 | 0.87 |
ENSDART00000185931
|
ndst3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr7_-_58130703 | 0.87 |
ENSDART00000172082
|
ank2b
|
ankyrin 2b, neuronal |
| chr5_-_8164439 | 0.86 |
ENSDART00000189912
|
slc1a3a
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3a |
| chr14_+_11458044 | 0.85 |
ENSDART00000186425
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr1_+_31942961 | 0.85 |
ENSDART00000007522
|
anos1a
|
anosmin 1a |
| chr5_+_4806851 | 0.82 |
ENSDART00000067599
|
angptl2a
|
angiopoietin-like 2a |
| chr12_-_11570 | 0.82 |
ENSDART00000186179
|
shisa6
|
shisa family member 6 |
| chr18_-_1228688 | 0.81 |
ENSDART00000064403
|
nptnb
|
neuroplastin b |
| chr5_-_63515210 | 0.81 |
ENSDART00000022348
|
prdm12b
|
PR domain containing 12b |
| chr7_+_6652967 | 0.79 |
ENSDART00000102681
|
pnp5a
|
purine nucleoside phosphorylase 5a |
| chr12_-_4756478 | 0.78 |
ENSDART00000152181
|
mapta
|
microtubule-associated protein tau a |
| chr19_-_2317558 | 0.78 |
ENSDART00000190300
|
sp8a
|
sp8 transcription factor a |
| chr4_+_76608282 | 0.77 |
ENSDART00000146610
|
ms4a17a.8
|
membrane-spanning 4-domains, subfamily A, member 17A.8 |
| chr10_-_22803740 | 0.77 |
ENSDART00000079469
ENSDART00000187968 ENSDART00000122543 |
pcolcea
|
procollagen C-endopeptidase enhancer a |
| chr8_-_1051438 | 0.76 |
ENSDART00000067093
ENSDART00000170737 |
smyd1b
|
SET and MYND domain containing 1b |
| chr14_+_11457500 | 0.75 |
ENSDART00000169202
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr5_+_250078 | 0.74 |
ENSDART00000127504
|
trabd2a
|
TraB domain containing 2A |
| chr3_-_36260102 | 0.74 |
ENSDART00000126588
|
rac3a
|
Rac family small GTPase 3a |
| chr16_-_14074594 | 0.72 |
ENSDART00000090234
|
trim109
|
tripartite motif containing 109 |
| chr6_-_21492752 | 0.72 |
ENSDART00000006843
ENSDART00000171479 |
cacng1a
|
calcium channel, voltage-dependent, gamma subunit 1a |
| chr25_-_23526058 | 0.72 |
ENSDART00000191331
ENSDART00000062930 |
phlda2
|
pleckstrin homology-like domain, family A, member 2 |
| chr17_+_27434626 | 0.71 |
ENSDART00000052446
|
vgll2b
|
vestigial-like family member 2b |
| chr17_+_52822422 | 0.70 |
ENSDART00000158273
ENSDART00000161414 |
meis2a
|
Meis homeobox 2a |
| chr19_-_2420990 | 0.70 |
ENSDART00000181498
|
TMEM196 (1 of many)
|
transmembrane protein 196 |
| chr23_+_23658474 | 0.70 |
ENSDART00000162838
|
agrn
|
agrin |
| chr7_-_69636502 | 0.69 |
ENSDART00000126739
|
tspan5a
|
tetraspanin 5a |
| chr16_-_42894628 | 0.69 |
ENSDART00000045600
|
hfe2
|
hemochromatosis type 2 |
| chr24_-_33703504 | 0.69 |
ENSDART00000079292
|
cavin4b
|
caveolae associated protein 4b |
| chr14_-_9281232 | 0.69 |
ENSDART00000054693
|
asb12b
|
ankyrin repeat and SOCS box-containing 12b |
| chr11_-_3343463 | 0.69 |
ENSDART00000066177
|
tuba2
|
tubulin, alpha 2 |
| chr23_-_46201008 | 0.68 |
ENSDART00000160110
|
tgm1l4
|
transglutaminase 1 like 4 |
| chr9_-_100579 | 0.68 |
ENSDART00000006099
|
lims2
|
LIM and senescent cell antigen-like domains 2 |
| chr4_-_18954001 | 0.68 |
ENSDART00000144814
|
si:dkey-31f5.8
|
si:dkey-31f5.8 |
| chr10_+_35265083 | 0.67 |
ENSDART00000048831
|
tmem120a
|
transmembrane protein 120A |
| chr23_-_15284757 | 0.65 |
ENSDART00000139135
|
sulf2b
|
sulfatase 2b |
| chr11_+_36477481 | 0.64 |
ENSDART00000128245
|
ldlrad2
|
low density lipoprotein receptor class A domain containing 2 |
| chr14_+_7939398 | 0.64 |
ENSDART00000189773
|
cxxc5b
|
CXXC finger protein 5b |
| chr5_-_26834511 | 0.63 |
ENSDART00000136713
ENSDART00000192932 ENSDART00000113246 |
si:ch211-102c2.4
|
si:ch211-102c2.4 |
| chr17_+_52823015 | 0.62 |
ENSDART00000160507
ENSDART00000186979 |
meis2a
|
Meis homeobox 2a |
| chr15_-_39969988 | 0.62 |
ENSDART00000146054
|
rps5
|
ribosomal protein S5 |
| chr14_-_32258759 | 0.61 |
ENSDART00000052949
|
fgf13a
|
fibroblast growth factor 13a |
| chr16_-_31188715 | 0.61 |
ENSDART00000058829
|
scrt1b
|
scratch family zinc finger 1b |
| chr8_-_4596662 | 0.61 |
ENSDART00000138199
|
sept5a
|
septin 5a |
| chr15_-_2652640 | 0.61 |
ENSDART00000146094
|
cldnf
|
claudin f |
| chr12_+_47663419 | 0.61 |
ENSDART00000171932
|
HHEX
|
hematopoietically expressed homeobox |
| chr12_-_656540 | 0.60 |
ENSDART00000172651
|
sult2st2
|
sulfotransferase family 2, cytosolic sulfotransferase 2 |
| chr14_-_36862745 | 0.59 |
ENSDART00000109293
|
rnf130
|
ring finger protein 130 |
| chr23_+_37458602 | 0.58 |
ENSDART00000181686
|
cdaa
|
cytidine deaminase a |
| chr13_-_7766758 | 0.58 |
ENSDART00000171831
|
h2afy2
|
H2A histone family, member Y2 |
| chr5_+_7564644 | 0.58 |
ENSDART00000192173
|
CABZ01039096.1
|
|
| chr20_+_19066596 | 0.57 |
ENSDART00000130271
|
sox7
|
SRY (sex determining region Y)-box 7 |
| chr15_-_18176694 | 0.57 |
ENSDART00000189840
|
tmprss5
|
transmembrane protease, serine 5 |
| chr23_-_3408777 | 0.56 |
ENSDART00000193245
|
mafba
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Ba |
| chr1_-_471925 | 0.56 |
ENSDART00000152684
|
zgc:92518
|
zgc:92518 |
| chr7_+_31838320 | 0.56 |
ENSDART00000144679
ENSDART00000174217 ENSDART00000122506 |
mybpc3
|
myosin binding protein C, cardiac |
| chr13_+_30903816 | 0.55 |
ENSDART00000191727
|
ercc6
|
excision repair cross-complementation group 6 |
| chr17_-_4318393 | 0.55 |
ENSDART00000167995
ENSDART00000153824 |
napba
|
N-ethylmaleimide-sensitive factor attachment protein, beta a |
| chr1_+_37195919 | 0.55 |
ENSDART00000159684
ENSDART00000172742 ENSDART00000158395 |
dclk2a
|
doublecortin-like kinase 2a |
| chr11_+_14622379 | 0.54 |
ENSDART00000112589
|
efna2b
|
ephrin-A2b |
| chr16_-_9980402 | 0.54 |
ENSDART00000066372
|
id4
|
inhibitor of DNA binding 4 |
| chr20_-_36887176 | 0.52 |
ENSDART00000143515
ENSDART00000146994 |
txlnba
|
taxilin beta a |
| chr13_-_7767044 | 0.52 |
ENSDART00000159453
|
h2afy2
|
H2A histone family, member Y2 |
| chr13_+_51579851 | 0.52 |
ENSDART00000163847
|
nkx6.2
|
NK6 homeobox 2 |
| chr1_-_206208 | 0.51 |
ENSDART00000060968
|
adprhl1
|
ADP-ribosylhydrolase like 1 |
| chr14_-_41678749 | 0.51 |
ENSDART00000163039
|
fgfrl1b
|
fibroblast growth factor receptor like 1b |
| chr13_-_27916439 | 0.51 |
ENSDART00000139081
ENSDART00000087097 |
ogfrl1
|
opioid growth factor receptor-like 1 |
| chr3_-_36115339 | 0.50 |
ENSDART00000187406
ENSDART00000123505 ENSDART00000151775 |
rab11fip4a
|
RAB11 family interacting protein 4 (class II) a |
| chr1_+_37196106 | 0.50 |
ENSDART00000008756
ENSDART00000157503 ENSDART00000162971 ENSDART00000191004 ENSDART00000078206 ENSDART00000045111 |
dclk2a
|
doublecortin-like kinase 2a |
| chr14_+_7939216 | 0.50 |
ENSDART00000171657
|
cxxc5b
|
CXXC finger protein 5b |
| chr6_+_16031189 | 0.50 |
ENSDART00000015333
|
gbx2
|
gastrulation brain homeobox 2 |
| chr20_-_36887516 | 0.50 |
ENSDART00000076313
|
txlnba
|
taxilin beta a |
| chr20_-_40319890 | 0.50 |
ENSDART00000075112
|
clvs2
|
clavesin 2 |
| chr12_+_6002715 | 0.50 |
ENSDART00000114961
|
si:ch211-131k2.3
|
si:ch211-131k2.3 |
| chr25_-_19433244 | 0.50 |
ENSDART00000154778
|
map1ab
|
microtubule-associated protein 1Ab |
| chr7_+_34231782 | 0.50 |
ENSDART00000173547
|
lctla
|
lactase-like a |
| chr23_-_30781875 | 0.49 |
ENSDART00000114628
ENSDART00000180949 ENSDART00000191313 |
myt1a
|
myelin transcription factor 1a |
| chr6_-_10964083 | 0.49 |
ENSDART00000181583
|
notum2
|
notum pectinacetylesterase 2 |
| chr6_-_609880 | 0.49 |
ENSDART00000149248
ENSDART00000148867 ENSDART00000149414 ENSDART00000148552 ENSDART00000148391 |
lgals2b
|
lectin, galactoside-binding, soluble, 2b |
| chr12_+_24562667 | 0.49 |
ENSDART00000056256
|
nrxn1a
|
neurexin 1a |
| chr19_-_46957968 | 0.48 |
ENSDART00000043713
|
angpt1
|
angiopoietin 1 |
| chr20_+_19066858 | 0.48 |
ENSDART00000192086
|
sox7
|
SRY (sex determining region Y)-box 7 |
| chr23_-_43609595 | 0.48 |
ENSDART00000172222
|
CABZ01117603.1
|
|
| chr16_-_13789908 | 0.48 |
ENSDART00000138540
|
ttyh1
|
tweety family member 1 |
| chr6_+_21095918 | 0.48 |
ENSDART00000167225
|
spega
|
SPEG complex locus a |
| chr3_-_26109322 | 0.47 |
ENSDART00000113780
|
zgc:162612
|
zgc:162612 |
| chr10_+_15777064 | 0.46 |
ENSDART00000114483
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr19_-_2421793 | 0.46 |
ENSDART00000180238
|
TMEM196 (1 of many)
|
transmembrane protein 196 |
| chr23_+_20931030 | 0.46 |
ENSDART00000167014
|
pax7b
|
paired box 7b |
| chr4_+_10066840 | 0.46 |
ENSDART00000026492
|
flncb
|
filamin C, gamma b (actin binding protein 280) |
| chr11_+_27364338 | 0.46 |
ENSDART00000186759
|
fbln2
|
fibulin 2 |
| chr9_-_5046315 | 0.45 |
ENSDART00000179087
ENSDART00000109954 |
nr4a2a
|
nuclear receptor subfamily 4, group A, member 2a |
| chr10_+_15777258 | 0.45 |
ENSDART00000140511
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr10_-_5053589 | 0.45 |
ENSDART00000193579
|
tmem150c
|
transmembrane protein 150C |
| chr22_-_968484 | 0.45 |
ENSDART00000105895
|
cacna1sa
|
calcium channel, voltage-dependent, L type, alpha 1S subunit, a |
| chr25_+_31868268 | 0.44 |
ENSDART00000022325
|
parp16
|
poly (ADP-ribose) polymerase family, member 16 |
| chr5_-_13818833 | 0.44 |
ENSDART00000192198
|
add2
|
adducin 2 (beta) |
| chr8_+_38415374 | 0.44 |
ENSDART00000085395
|
nkx6.3
|
NK6 homeobox 3 |
| chr1_+_39553040 | 0.43 |
ENSDART00000137676
|
tenm3
|
teneurin transmembrane protein 3 |
| chr19_-_43657468 | 0.43 |
ENSDART00000150940
|
si:ch211-193k19.2
|
si:ch211-193k19.2 |
| chr5_+_42467867 | 0.43 |
ENSDART00000172028
|
pimr58
|
Pim proto-oncogene, serine/threonine kinase, related 58 |
| chr10_-_34741738 | 0.43 |
ENSDART00000163072
|
dclk1a
|
doublecortin-like kinase 1a |
| chr8_-_50259448 | 0.42 |
ENSDART00000146056
|
nkx3-1
|
NK3 homeobox 1 |
| chr19_+_1964005 | 0.42 |
ENSDART00000172049
|
sh3bp5a
|
SH3-domain binding protein 5a (BTK-associated) |
| chr23_-_44219902 | 0.42 |
ENSDART00000185874
|
zgc:158659
|
zgc:158659 |
| chr17_-_51195651 | 0.42 |
ENSDART00000191205
ENSDART00000088185 |
paplna
|
papilin a, proteoglycan-like sulfated glycoprotein |
| chr18_-_49020066 | 0.41 |
ENSDART00000174394
|
BX663503.3
|
|
| chr16_+_3004422 | 0.41 |
ENSDART00000189969
|
CABZ01043952.1
|
|
| chr24_+_39108243 | 0.41 |
ENSDART00000156353
|
mss51
|
MSS51 mitochondrial translational activator |
| chr9_-_48281941 | 0.41 |
ENSDART00000099787
|
klhl41a
|
kelch-like family member 41a |
| chr18_+_30508729 | 0.41 |
ENSDART00000185140
|
cox4i1
|
cytochrome c oxidase subunit IV isoform 1 |
| chr5_-_48307804 | 0.41 |
ENSDART00000182831
ENSDART00000186920 ENSDART00000183585 |
mef2cb
|
myocyte enhancer factor 2cb |
| chr9_-_68934 | 0.41 |
ENSDART00000054594
ENSDART00000009389 |
il10rb
|
interleukin 10 receptor, beta |
| chr25_+_13406069 | 0.41 |
ENSDART00000010495
|
znrf1
|
zinc and ring finger 1 |
| chr7_-_26924903 | 0.40 |
ENSDART00000124363
|
alx4a
|
ALX homeobox 4a |
| chr7_+_46003449 | 0.40 |
ENSDART00000159700
ENSDART00000173625 |
si:ch211-260e23.9
|
si:ch211-260e23.9 |
| chr3_-_51912019 | 0.40 |
ENSDART00000149914
|
aatka
|
apoptosis-associated tyrosine kinase a |
| chr16_+_41873708 | 0.40 |
ENSDART00000084631
ENSDART00000084639 ENSDART00000058611 |
scn1ba
|
sodium channel, voltage-gated, type I, beta a |
| chr8_+_54284961 | 0.39 |
ENSDART00000122692
|
plxnd1
|
plexin D1 |
| chr8_+_39570615 | 0.39 |
ENSDART00000142557
|
lzts1
|
leucine zipper, putative tumor suppressor 1 |
| chr4_+_16787040 | 0.39 |
ENSDART00000039027
|
golt1ba
|
golgi transport 1Ba |
| chr25_-_35960229 | 0.39 |
ENSDART00000073434
|
snx20
|
sorting nexin 20 |
| chr23_+_19977120 | 0.39 |
ENSDART00000089342
|
cfap126
|
cilia and flagella associated protein 126 |
| chr11_-_11471857 | 0.39 |
ENSDART00000030103
|
krt94
|
keratin 94 |
| chr11_+_7264457 | 0.39 |
ENSDART00000154182
|
reep6
|
receptor accessory protein 6 |
| chr12_-_17201028 | 0.39 |
ENSDART00000020541
|
lipf
|
lipase, gastric |
| chr8_-_26609259 | 0.38 |
ENSDART00000027301
|
sema3b
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B |
| chr24_+_6107901 | 0.38 |
ENSDART00000156419
|
si:ch211-37e10.2
|
si:ch211-37e10.2 |
| chr16_+_43344475 | 0.38 |
ENSDART00000085282
|
mrpl32
|
mitochondrial ribosomal protein L32 |
| chr3_+_24458899 | 0.38 |
ENSDART00000156655
|
cbx6b
|
chromobox homolog 6b |
| chr7_+_35229645 | 0.37 |
ENSDART00000144327
|
tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr13_-_11072964 | 0.37 |
ENSDART00000135989
|
cep170aa
|
centrosomal protein 170Aa |
| chr1_+_50987535 | 0.37 |
ENSDART00000140657
|
mdh1aa
|
malate dehydrogenase 1Aa, NAD (soluble) |
| chr17_-_10073926 | 0.37 |
ENSDART00000166081
ENSDART00000161574 |
zgc:109986
|
zgc:109986 |
| chr11_+_21050326 | 0.36 |
ENSDART00000065984
|
zgc:113307
|
zgc:113307 |
| chr19_-_30904590 | 0.36 |
ENSDART00000137633
|
si:ch211-194e15.5
|
si:ch211-194e15.5 |
| chr14_-_29906209 | 0.36 |
ENSDART00000192952
|
sorbs2b
|
sorbin and SH3 domain containing 2b |
| chr23_-_31266586 | 0.36 |
ENSDART00000139746
|
si:dkey-261l7.2
|
si:dkey-261l7.2 |
| chr19_+_9174166 | 0.36 |
ENSDART00000104637
ENSDART00000150968 |
si:ch211-81a5.8
|
si:ch211-81a5.8 |
| chr4_+_16787488 | 0.35 |
ENSDART00000143006
|
golt1ba
|
golgi transport 1Ba |
| chr3_+_18097700 | 0.35 |
ENSDART00000021634
|
wfikkn2a
|
info WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2a |
| chr9_+_34127005 | 0.34 |
ENSDART00000167384
ENSDART00000078065 |
f5
|
coagulation factor V |
| chr5_+_57320113 | 0.34 |
ENSDART00000036331
|
atp6v1g1
|
ATPase H+ transporting V1 subunit G1 |
| chr2_-_37888429 | 0.34 |
ENSDART00000183277
|
mbl2
|
mannose binding lectin 2 |
| chr20_-_36809059 | 0.34 |
ENSDART00000062925
|
slc25a27
|
solute carrier family 25, member 27 |
| chr9_-_18568927 | 0.34 |
ENSDART00000084668
|
enox1
|
ecto-NOX disulfide-thiol exchanger 1 |
| chr11_+_27364059 | 0.34 |
ENSDART00000172883
|
fbln2
|
fibulin 2 |
| chr12_+_17504559 | 0.34 |
ENSDART00000020628
|
cyth3a
|
cytohesin 3a |
| chr4_+_3980247 | 0.33 |
ENSDART00000049194
|
gpr37b
|
G protein-coupled receptor 37b |
| chr18_+_36631923 | 0.33 |
ENSDART00000098980
|
znf296
|
zinc finger protein 296 |
| chr9_-_327901 | 0.33 |
ENSDART00000159956
|
ndufa4l2a
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2a |
| chr23_+_43177290 | 0.33 |
ENSDART00000193300
ENSDART00000186065 |
si:dkey-65j6.2
|
si:dkey-65j6.2 |
| chr17_-_7861219 | 0.33 |
ENSDART00000148604
|
syne1b
|
spectrin repeat containing, nuclear envelope 1b |
| chr5_-_39474235 | 0.33 |
ENSDART00000171557
|
antxr2a
|
anthrax toxin receptor 2a |
Network of associatons between targets according to the STRING database.
First level regulatory network of myog
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.4 | GO:0090076 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) negative regulation of striated muscle contraction(GO:0045988) relaxation of skeletal muscle(GO:0090076) |
| 0.7 | 2.1 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.6 | 1.9 | GO:0006600 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 0.4 | 1.4 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.3 | 1.6 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.2 | 0.7 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.2 | 0.9 | GO:0048521 | positive regulation of epithelial cell differentiation(GO:0030858) negative regulation of behavior(GO:0048521) negative regulation of feeding behavior(GO:2000252) |
| 0.2 | 0.9 | GO:0014857 | skeletal muscle cell proliferation(GO:0014856) regulation of skeletal muscle cell proliferation(GO:0014857) |
| 0.2 | 1.4 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.2 | 1.1 | GO:0006972 | hyperosmotic response(GO:0006972) |
| 0.2 | 1.8 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.2 | 1.2 | GO:0035989 | tendon development(GO:0035989) |
| 0.2 | 0.6 | GO:0009972 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.2 | 0.9 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.2 | 0.5 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.2 | 0.5 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.2 | 0.9 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.2 | 1.1 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.1 | 0.7 | GO:1902514 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.1 | 0.5 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.1 | 1.0 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.1 | 0.5 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.1 | 0.5 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.1 | 0.4 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 0.6 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.1 | 0.6 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.1 | 0.4 | GO:1903385 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 0.3 | GO:0015889 | cobalt ion transport(GO:0006824) cobalamin transport(GO:0015889) |
| 0.1 | 1.9 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 0.5 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.1 | 0.5 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 0.1 | 0.8 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 0.7 | GO:0033292 | T-tubule organization(GO:0033292) |
| 0.1 | 0.3 | GO:2000008 | regulation of protein localization to cell surface(GO:2000008) negative regulation of protein localization to cell surface(GO:2000009) |
| 0.1 | 0.7 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 3.3 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 0.9 | GO:0060753 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.1 | 0.3 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.1 | 0.4 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 0.9 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 0.6 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 | 1.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.2 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.1 | 0.8 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.1 | 1.2 | GO:0006942 | regulation of striated muscle contraction(GO:0006942) |
| 0.1 | 0.4 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.1 | 0.4 | GO:0086091 | regulation of heart rate by cardiac conduction(GO:0086091) |
| 0.1 | 0.3 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.1 | 0.7 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.1 | 1.5 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.1 | 0.2 | GO:0099623 | regulation of membrane repolarization(GO:0060306) regulation of ventricular cardiac muscle cell membrane repolarization(GO:0060307) regulation of cardiac muscle cell membrane repolarization(GO:0099623) ventricular cardiac muscle cell membrane repolarization(GO:0099625) |
| 0.1 | 0.2 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.1 | 0.2 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.1 | 0.2 | GO:0042543 | protein N-linked glycosylation via arginine(GO:0042543) |
| 0.0 | 0.6 | GO:0035860 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.5 | GO:0003307 | regulation of Wnt signaling pathway involved in heart development(GO:0003307) |
| 0.0 | 0.4 | GO:0031498 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.7 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.4 | GO:0021871 | forebrain regionalization(GO:0021871) |
| 0.0 | 0.2 | GO:0090155 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.0 | 0.7 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.1 | GO:0097066 | response to thyroid hormone(GO:0097066) |
| 0.0 | 0.3 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.1 | GO:0008344 | adult locomotory behavior(GO:0008344) |
| 0.0 | 0.4 | GO:0050732 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.1 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.0 | 0.5 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 0.1 | GO:0014014 | negative regulation of gliogenesis(GO:0014014) |
| 0.0 | 1.0 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.5 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.2 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.7 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.1 | GO:0008584 | male gonad development(GO:0008584) |
| 0.0 | 0.3 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.1 | GO:0048730 | epidermis morphogenesis(GO:0048730) |
| 0.0 | 0.3 | GO:0021703 | locus ceruleus development(GO:0021703) |
| 0.0 | 0.6 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.2 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.0 | 0.3 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 1.1 | GO:0060840 | artery development(GO:0060840) |
| 0.0 | 0.4 | GO:0035476 | angioblast cell migration(GO:0035476) |
| 0.0 | 0.1 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 1.3 | GO:0021575 | hindbrain morphogenesis(GO:0021575) |
| 0.0 | 0.7 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.2 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.1 | GO:0055107 | Golgi to secretory granule transport(GO:0055107) |
| 0.0 | 0.5 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.1 | GO:0035889 | otolith tethering(GO:0035889) |
| 0.0 | 0.3 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 0.2 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.6 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.1 | GO:0014896 | muscle hypertrophy(GO:0014896) |
| 0.0 | 0.1 | GO:1903817 | negative regulation of potassium ion transmembrane transporter activity(GO:1901017) negative regulation of potassium ion transmembrane transport(GO:1901380) negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 0.5 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.7 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.0 | 0.2 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.1 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.1 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 1.2 | GO:0009142 | nucleoside triphosphate biosynthetic process(GO:0009142) |
| 0.0 | 1.7 | GO:0061515 | myeloid cell development(GO:0061515) |
| 0.0 | 0.3 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.0 | 0.5 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.3 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.1 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.0 | 0.4 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.0 | 0.1 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.0 | 0.3 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.1 | GO:0016037 | light absorption(GO:0016037) |
| 0.0 | 0.1 | GO:0070293 | renal absorption(GO:0070293) |
| 0.0 | 0.3 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.1 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.2 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.3 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.5 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 0.5 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.1 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.2 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.2 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.0 | 0.2 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.1 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.0 | 0.3 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 1.7 | GO:0048703 | embryonic viscerocranium morphogenesis(GO:0048703) |
| 0.0 | 0.2 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.0 | 0.4 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.0 | 0.1 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.2 | GO:1900153 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 | 0.1 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.2 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 0.5 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 1.1 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.0 | GO:0044406 | virion attachment to host cell(GO:0019062) adhesion of symbiont to host(GO:0044406) adhesion of symbiont to host cell(GO:0044650) |
| 0.0 | 0.5 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.3 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 1.1 | GO:0006909 | phagocytosis(GO:0006909) |
| 0.0 | 0.2 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.0 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 1.3 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.4 | GO:0031673 | H zone(GO:0031673) |
| 0.2 | 0.5 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 0.5 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 1.0 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 3.0 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 1.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.7 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 0.9 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 1.1 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 0.3 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.1 | 3.4 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 1.6 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 0.4 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.7 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.8 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.1 | GO:0097541 | axonemal basal plate(GO:0097541) |
| 0.0 | 1.0 | GO:0016528 | sarcoplasm(GO:0016528) sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.2 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 0.9 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.2 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.3 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.4 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.2 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.0 | 0.3 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 1.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.3 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.4 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 1.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.3 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.1 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.5 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.2 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.4 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 1.6 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.1 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.0 | 0.3 | GO:0032589 | neuron projection membrane(GO:0032589) dendrite membrane(GO:0032590) |
| 0.0 | 0.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.6 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.9 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 1.1 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 0.1 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.7 | GO:0005604 | basement membrane(GO:0005604) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0071253 | connexin binding(GO:0071253) |
| 0.3 | 0.8 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.2 | 0.9 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.2 | 3.4 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.2 | 0.5 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.1 | 0.6 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.1 | 0.4 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.1 | 1.0 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.5 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.1 | 0.9 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 1.2 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.1 | 1.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.5 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 0.3 | GO:0004776 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.1 | 0.8 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 0.6 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 1.0 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 0.4 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.1 | 0.6 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 1.0 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 0.2 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 0.5 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 0.2 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 1.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 1.2 | GO:0043236 | laminin binding(GO:0043236) |
| 0.1 | 0.6 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.3 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.9 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.0 | 1.2 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.3 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.5 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.3 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.5 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.9 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.9 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.6 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.5 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.7 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.3 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.9 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 1.5 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.3 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.3 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.1 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.0 | 0.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.0 | 0.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.7 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.3 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.2 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.1 | GO:0000810 | diacylglycerol diphosphate phosphatase activity(GO:0000810) |
| 0.0 | 0.7 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.1 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.0 | 0.8 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.7 | GO:0098632 | protein binding involved in cell adhesion(GO:0098631) protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.3 | GO:0022884 | macromolecule transmembrane transporter activity(GO:0022884) |
| 0.0 | 0.5 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.1 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.0 | 0.2 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.3 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.0 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 2.7 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.2 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 1.0 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.2 | GO:0005165 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.0 | 0.4 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.5 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.0 | 0.2 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.4 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.1 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.0 | 0.2 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.3 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.1 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.3 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.0 | 0.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.2 | GO:0019209 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.2 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.9 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.6 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.7 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.5 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.2 | 1.9 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 1.3 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 1.6 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 0.9 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.4 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.6 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 0.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.7 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.6 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.1 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |