Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
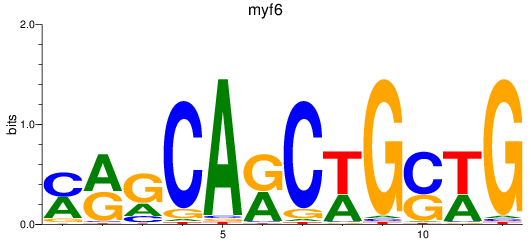
Results for myf6
Z-value: 1.05
Transcription factors associated with myf6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
myf6
|
ENSDARG00000029830 | myogenic factor 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| myf6 | dr11_v1_chr4_+_21717793_21717793 | -0.79 | 1.0e-02 | Click! |
Activity profile of myf6 motif
Sorted Z-values of myf6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_+_2714949 | 1.23 |
ENSDART00000105284
|
ncoa6
|
nuclear receptor coactivator 6 |
| chr17_-_51818659 | 1.04 |
ENSDART00000111389
ENSDART00000157244 |
exd2
|
exonuclease 3'-5' domain containing 2 |
| chr25_+_34889061 | 0.98 |
ENSDART00000136226
|
spire2
|
spire-type actin nucleation factor 2 |
| chr6_+_154556 | 0.96 |
ENSDART00000193153
|
zglp1
|
zinc finger, GATA-like protein 1 |
| chr9_+_22657221 | 0.95 |
ENSDART00000101765
|
si:dkey-189g17.2
|
si:dkey-189g17.2 |
| chr22_-_37738203 | 0.93 |
ENSDART00000143190
|
acap2
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 |
| chr19_-_18313303 | 0.81 |
ENSDART00000164644
ENSDART00000167480 ENSDART00000163104 |
si:dkey-208k4.2
|
si:dkey-208k4.2 |
| chr2_+_27855102 | 0.79 |
ENSDART00000150330
|
buc
|
bucky ball |
| chr7_-_20582842 | 0.76 |
ENSDART00000169750
ENSDART00000111719 |
si:dkey-19b23.11
|
si:dkey-19b23.11 |
| chr2_+_27855346 | 0.76 |
ENSDART00000175159
ENSDART00000192645 |
buc
|
bucky ball |
| chr9_+_22656976 | 0.75 |
ENSDART00000136249
ENSDART00000139270 |
si:dkey-189g17.2
|
si:dkey-189g17.2 |
| chr19_+_41479990 | 0.74 |
ENSDART00000087187
|
ago2
|
argonaute RISC catalytic component 2 |
| chr6_+_153146 | 0.72 |
ENSDART00000097468
|
zglp1
|
zinc finger, GATA-like protein 1 |
| chr25_+_34888886 | 0.72 |
ENSDART00000035245
|
spire2
|
spire-type actin nucleation factor 2 |
| chr3_-_2613990 | 0.68 |
ENSDART00000137102
|
si:dkey-217f16.6
|
si:dkey-217f16.6 |
| chr12_-_10512911 | 0.68 |
ENSDART00000124562
ENSDART00000106163 |
zgc:152977
|
zgc:152977 |
| chr14_-_7409364 | 0.67 |
ENSDART00000036463
|
dnd1
|
DND microRNA-mediated repression inhibitor 1 |
| chr22_+_25274712 | 0.66 |
ENSDART00000137341
|
BX950205.1
|
|
| chr16_+_53278406 | 0.65 |
ENSDART00000010792
|
ptdss1a
|
phosphatidylserine synthase 1a |
| chr2_+_6253246 | 0.64 |
ENSDART00000058256
ENSDART00000076700 |
zp3b
|
zona pellucida glycoprotein 3b |
| chr5_-_33230794 | 0.64 |
ENSDART00000144273
|
kank1b
|
KN motif and ankyrin repeat domains 1b |
| chr10_-_24784446 | 0.64 |
ENSDART00000140877
|
si:ch1073-15f19.2
|
si:ch1073-15f19.2 |
| chr23_+_37579107 | 0.63 |
ENSDART00000169376
|
plekhg5b
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5b |
| chr8_-_20138054 | 0.63 |
ENSDART00000133141
ENSDART00000147634 ENSDART00000029939 |
rfx2
|
regulatory factor X, 2 (influences HLA class II expression) |
| chr14_+_28518349 | 0.62 |
ENSDART00000159961
|
stag2b
|
stromal antigen 2b |
| chr9_-_105135 | 0.62 |
ENSDART00000180126
|
FQ377903.3
|
|
| chr22_-_5099824 | 0.60 |
ENSDART00000122341
ENSDART00000161345 |
zfr2
|
zinc finger RNA binding protein 2 |
| chr8_+_3434146 | 0.60 |
ENSDART00000164426
|
ctu1
|
cytosolic thiouridylase subunit 1 homolog (S. pombe) |
| chr19_-_10207103 | 0.59 |
ENSDART00000151629
|
znf865
|
zinc finger protein 865 |
| chr23_+_45845159 | 0.58 |
ENSDART00000023944
|
lmnl3
|
lamin L3 |
| chr16_+_48729947 | 0.58 |
ENSDART00000049051
|
brd2b
|
bromodomain containing 2b |
| chr23_+_43668756 | 0.57 |
ENSDART00000112598
|
otud4
|
OTU deubiquitinase 4 |
| chr12_+_20693743 | 0.56 |
ENSDART00000153023
ENSDART00000153370 |
st6galnac1.2
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1, tandem duplicate 2 |
| chr20_-_9199721 | 0.56 |
ENSDART00000064140
|
ylpm1
|
YLP motif containing 1 |
| chr7_-_59589547 | 0.55 |
ENSDART00000008554
ENSDART00000101731 |
larp7
|
La ribonucleoprotein domain family, member 7 |
| chr19_+_14454306 | 0.54 |
ENSDART00000161965
|
zdhhc18b
|
zinc finger, DHHC-type containing 18b |
| chr2_+_35116627 | 0.54 |
ENSDART00000113609
|
cop1
|
COP1, E3 ubiquitin ligase |
| chr11_+_11120532 | 0.53 |
ENSDART00000026135
ENSDART00000189872 |
ly75
|
lymphocyte antigen 75 |
| chr21_-_38717854 | 0.52 |
ENSDART00000065169
ENSDART00000113813 |
siah2l
|
seven in absentia homolog 2 (Drosophila)-like |
| chr24_+_39034090 | 0.52 |
ENSDART00000185763
|
capn15
|
calpain 15 |
| chr14_+_30774032 | 0.52 |
ENSDART00000139552
|
atl3
|
atlastin 3 |
| chr5_+_13521081 | 0.51 |
ENSDART00000171975
|
si:ch211-230g14.6
|
si:ch211-230g14.6 |
| chr11_-_34480822 | 0.51 |
ENSDART00000129029
|
xxylt1
|
xyloside xylosyltransferase 1 |
| chr12_-_10508952 | 0.51 |
ENSDART00000152806
|
zgc:152977
|
zgc:152977 |
| chr21_+_43199237 | 0.51 |
ENSDART00000151748
|
aff4
|
AF4/FMR2 family, member 4 |
| chr4_-_22335247 | 0.51 |
ENSDART00000192781
|
golgb1
|
golgin B1 |
| chr23_+_27782071 | 0.49 |
ENSDART00000131379
|
kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr19_-_30810328 | 0.49 |
ENSDART00000184875
|
myclb
|
MYCL proto-oncogene, bHLH transcription factor b |
| chr2_-_57707039 | 0.49 |
ENSDART00000097685
|
CABZ01060891.1
|
|
| chr20_-_38787047 | 0.48 |
ENSDART00000152913
ENSDART00000153430 |
dnajc5ga
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma a |
| chr3_-_15679107 | 0.48 |
ENSDART00000080441
|
zgc:66443
|
zgc:66443 |
| chr2_+_44518636 | 0.48 |
ENSDART00000153733
|
pask
|
PAS domain containing serine/threonine kinase |
| chr18_-_25771553 | 0.47 |
ENSDART00000103046
|
zgc:162879
|
zgc:162879 |
| chr18_+_14277003 | 0.47 |
ENSDART00000006628
|
zgc:173742
|
zgc:173742 |
| chr24_-_1657276 | 0.47 |
ENSDART00000168131
|
si:ch73-378g22.1
|
si:ch73-378g22.1 |
| chr8_+_53344726 | 0.46 |
ENSDART00000184395
ENSDART00000170212 |
CU914536.1
|
|
| chr3_+_22935183 | 0.46 |
ENSDART00000157378
|
hdac5
|
histone deacetylase 5 |
| chr12_+_9849476 | 0.46 |
ENSDART00000110458
|
fam117ab
|
family with sequence similarity 117, member Ab |
| chr17_+_24597001 | 0.46 |
ENSDART00000191834
|
rlf
|
rearranged L-myc fusion |
| chr13_-_4848889 | 0.45 |
ENSDART00000165259
|
mcu
|
mitochondrial calcium uniporter |
| chr20_+_2642855 | 0.45 |
ENSDART00000058775
|
zgc:101562
|
zgc:101562 |
| chr20_-_49889111 | 0.45 |
ENSDART00000058858
|
kif13bb
|
kinesin family member 13Bb |
| chr23_+_19790962 | 0.45 |
ENSDART00000142228
|
flna
|
filamin A, alpha (actin binding protein 280) |
| chr21_+_33249478 | 0.45 |
ENSDART00000169972
|
si:ch211-151g22.1
|
si:ch211-151g22.1 |
| chr16_+_26601364 | 0.45 |
ENSDART00000087537
|
epb41l4b
|
erythrocyte membrane protein band 4.1 like 4B |
| chr5_-_9678075 | 0.44 |
ENSDART00000097217
|
FAM109B
|
si:ch211-193c2.2 |
| chr23_-_45319592 | 0.44 |
ENSDART00000189861
|
ccdc171
|
coiled-coil domain containing 171 |
| chr16_+_39146696 | 0.44 |
ENSDART00000121756
ENSDART00000084381 |
sybu
|
syntabulin (syntaxin-interacting) |
| chr24_+_41989108 | 0.44 |
ENSDART00000169725
|
zbtb14
|
zinc finger and BTB domain containing 14 |
| chr22_+_25352530 | 0.44 |
ENSDART00000176742
ENSDART00000113186 |
si:dkey-240e12.6
|
si:dkey-240e12.6 |
| chr13_-_4986364 | 0.44 |
ENSDART00000168465
|
nolc1
|
nucleolar and coiled-body phosphoprotein 1 |
| chr14_-_34605607 | 0.44 |
ENSDART00000191608
|
afap1l1a
|
actin filament associated protein 1-like 1a |
| chr11_-_40257225 | 0.43 |
ENSDART00000139009
|
TRIM62 (1 of many)
|
si:ch211-193i15.2 |
| chr8_-_22326744 | 0.43 |
ENSDART00000137645
|
cep104
|
centrosomal protein 104 |
| chr7_-_22632690 | 0.43 |
ENSDART00000165245
|
si:dkey-112a7.4
|
si:dkey-112a7.4 |
| chr3_+_32577994 | 0.43 |
ENSDART00000180643
|
si:ch73-248e21.1
|
si:ch73-248e21.1 |
| chr9_-_19366538 | 0.43 |
ENSDART00000138431
|
zgc:152951
|
zgc:152951 |
| chr24_-_20641000 | 0.43 |
ENSDART00000166135
|
zbtb47b
|
zinc finger and BTB domain containing 47b |
| chr18_+_6641542 | 0.43 |
ENSDART00000160379
|
c2cd5
|
C2 calcium-dependent domain containing 5 |
| chr23_+_45845423 | 0.43 |
ENSDART00000183404
|
lmnl3
|
lamin L3 |
| chr12_+_30788912 | 0.42 |
ENSDART00000160422
|
aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr5_-_30074332 | 0.42 |
ENSDART00000147963
|
bco2a
|
beta-carotene oxygenase 2a |
| chr19_+_7636941 | 0.42 |
ENSDART00000081611
ENSDART00000163805 ENSDART00000112404 |
cgnb
|
cingulin b |
| chr12_+_46582727 | 0.42 |
ENSDART00000149326
|
ush1gb
|
Usher syndrome 1Gb (autosomal recessive) |
| chr20_+_52774730 | 0.42 |
ENSDART00000014606
|
phactr1
|
phosphatase and actin regulator 1 |
| chr18_+_36786842 | 0.41 |
ENSDART00000123264
|
si:ch211-160d20.3
|
si:ch211-160d20.3 |
| chr6_-_2134581 | 0.41 |
ENSDART00000175478
|
vstm2l
|
V-set and transmembrane domain containing 2 like |
| chr11_+_6882362 | 0.41 |
ENSDART00000144181
|
klhl26
|
kelch-like family member 26 |
| chr9_-_41025062 | 0.41 |
ENSDART00000002053
|
pms1
|
PMS1 homolog 1, mismatch repair system component |
| chr23_-_27045231 | 0.41 |
ENSDART00000187979
|
zgc:66440
|
zgc:66440 |
| chr21_+_34167178 | 0.41 |
ENSDART00000158308
|
trpc5b
|
transient receptor potential cation channel, subfamily C, member 5b |
| chr18_-_43866001 | 0.41 |
ENSDART00000150218
|
treh
|
trehalase (brush-border membrane glycoprotein) |
| chr21_+_4204860 | 0.41 |
ENSDART00000146541
|
rapgef1b
|
Rap guanine nucleotide exchange factor (GEF) 1b |
| chr9_+_28232522 | 0.41 |
ENSDART00000031761
|
fzd5
|
frizzled class receptor 5 |
| chr1_+_218524 | 0.41 |
ENSDART00000109529
|
tmco3
|
transmembrane and coiled-coil domains 3 |
| chr5_+_36781732 | 0.41 |
ENSDART00000087191
|
mark4a
|
MAP/microtubule affinity-regulating kinase 4a |
| chr11_+_45448212 | 0.41 |
ENSDART00000173341
|
sos1
|
son of sevenless homolog 1 (Drosophila) |
| chr25_+_2776511 | 0.41 |
ENSDART00000115280
|
neo1b
|
neogenin 1b |
| chr14_-_34605804 | 0.41 |
ENSDART00000144547
|
afap1l1a
|
actin filament associated protein 1-like 1a |
| chr4_+_279669 | 0.40 |
ENSDART00000184884
|
CABZ01085275.1
|
|
| chr15_+_14592624 | 0.40 |
ENSDART00000162350
|
FBXO46
|
si:dkey-114g7.4 |
| chr11_+_6010177 | 0.40 |
ENSDART00000170047
ENSDART00000022526 ENSDART00000161001 ENSDART00000188999 |
gtpbp3
|
GTP binding protein 3, mitochondrial |
| chr22_+_25236657 | 0.40 |
ENSDART00000138012
|
zgc:172218
|
zgc:172218 |
| chr2_-_38114370 | 0.40 |
ENSDART00000131837
|
chd8
|
chromodomain helicase DNA binding protein 8 |
| chr3_-_31879201 | 0.40 |
ENSDART00000076189
|
zgc:171779
|
zgc:171779 |
| chr21_-_43636595 | 0.40 |
ENSDART00000151115
ENSDART00000151486 ENSDART00000151778 |
si:ch1073-263o8.2
|
si:ch1073-263o8.2 |
| chr7_-_61279523 | 0.40 |
ENSDART00000098610
|
rell1
|
RELT like 1 |
| chr8_-_22539113 | 0.40 |
ENSDART00000183297
ENSDART00000185981 |
csde1
|
cold shock domain containing E1, RNA-binding |
| chr15_-_7337148 | 0.39 |
ENSDART00000182568
|
SLC7A1 (1 of many)
|
high affinity cationic amino acid transporter 1 |
| chr15_-_7337537 | 0.39 |
ENSDART00000161613
|
SLC7A1 (1 of many)
|
high affinity cationic amino acid transporter 1 |
| chr7_+_24729558 | 0.39 |
ENSDART00000111542
ENSDART00000170100 |
shroom4
|
shroom family member 4 |
| chr15_+_30158652 | 0.39 |
ENSDART00000190682
|
nlk2
|
nemo-like kinase, type 2 |
| chr17_+_12075805 | 0.39 |
ENSDART00000155329
|
cnsta
|
consortin, connexin sorting protein a |
| chr2_-_57469115 | 0.39 |
ENSDART00000192201
|
pias4b
|
protein inhibitor of activated STAT, 4b |
| chr18_+_15876385 | 0.39 |
ENSDART00000142527
|
eea1
|
early endosome antigen 1 |
| chr13_+_28618086 | 0.39 |
ENSDART00000087001
|
cnnm2a
|
cyclin and CBS domain divalent metal cation transport mediator 2a |
| chr17_+_5931530 | 0.39 |
ENSDART00000168326
ENSDART00000189790 |
znf513b
|
zinc finger protein 513b |
| chr16_+_25011994 | 0.38 |
ENSDART00000157312
|
znf1035
|
zinc finger protein 1035 |
| chr1_-_9195629 | 0.38 |
ENSDART00000143587
ENSDART00000192174 |
ern2
|
endoplasmic reticulum to nucleus signaling 2 |
| chr7_+_24494299 | 0.38 |
ENSDART00000087568
|
nelfa
|
negative elongation factor complex member A |
| chr17_-_38887424 | 0.38 |
ENSDART00000141177
|
slc24a4a
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4a |
| chr10_-_8197049 | 0.38 |
ENSDART00000129467
|
dhx29
|
DEAH (Asp-Glu-Ala-His) box polypeptide 29 |
| chr13_-_12021566 | 0.38 |
ENSDART00000125430
|
pprc1
|
peroxisome proliferator-activated receptor gamma, coactivator-related 1 |
| chr4_-_20081621 | 0.38 |
ENSDART00000024647
|
dennd6b
|
DENN/MADD domain containing 6B |
| chr25_+_36328524 | 0.38 |
ENSDART00000073404
|
hist1h2a4
|
histone cluster 1 H2A family member 4 |
| chr8_+_13364950 | 0.37 |
ENSDART00000159760
|
slc5a5
|
solute carrier family 5 (sodium/iodide cotransporter), member 5 |
| chr24_-_5691956 | 0.37 |
ENSDART00000189112
|
dia1b
|
deleted in autism 1b |
| chr20_-_1265562 | 0.37 |
ENSDART00000189866
|
lats1
|
large tumor suppressor kinase 1 |
| chr3_-_11828206 | 0.37 |
ENSDART00000018159
|
si:ch211-262e15.1
|
si:ch211-262e15.1 |
| chr15_+_5116179 | 0.37 |
ENSDART00000101937
|
pgm2l1
|
phosphoglucomutase 2-like 1 |
| chr25_-_32869794 | 0.37 |
ENSDART00000162784
|
tmem266
|
transmembrane protein 266 |
| chr19_-_867071 | 0.37 |
ENSDART00000122257
|
eomesa
|
eomesodermin homolog a |
| chr5_-_18897482 | 0.36 |
ENSDART00000010101
ENSDART00000099434 |
aacs
|
acetoacetyl-CoA synthetase |
| chr20_+_34502606 | 0.36 |
ENSDART00000139739
|
gorab
|
golgin, rab6-interacting |
| chr3_-_27880229 | 0.36 |
ENSDART00000151404
|
abat
|
4-aminobutyrate aminotransferase |
| chr12_+_47794089 | 0.36 |
ENSDART00000160726
|
polr3a
|
polymerase (RNA) III (DNA directed) polypeptide A |
| chr4_-_8093753 | 0.36 |
ENSDART00000133434
|
wnk1b
|
WNK lysine deficient protein kinase 1b |
| chr6_-_39198912 | 0.36 |
ENSDART00000077938
|
c1galt1b
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1b |
| chr5_-_38384755 | 0.36 |
ENSDART00000188573
ENSDART00000051233 |
mink1
|
misshapen-like kinase 1 |
| chr23_+_6544453 | 0.36 |
ENSDART00000005373
|
spo11
|
SPO11 meiotic protein covalently bound to DSB |
| chr15_+_509126 | 0.36 |
ENSDART00000102274
|
ftr86
|
finTRIM family, member 86 |
| chr12_-_24832297 | 0.36 |
ENSDART00000066317
|
foxn2b
|
forkhead box N2b |
| chr5_+_37744625 | 0.36 |
ENSDART00000014031
|
dpf2
|
D4, zinc and double PHD fingers family 2 |
| chr22_-_16755885 | 0.35 |
ENSDART00000036467
|
patj
|
PATJ, crumbs cell polarity complex component |
| chr8_+_14915332 | 0.35 |
ENSDART00000164385
|
abl2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr2_-_24069331 | 0.35 |
ENSDART00000156972
ENSDART00000181691 ENSDART00000157041 |
slc12a7a
|
solute carrier family 12 (potassium/chloride transporter), member 7a |
| chr19_-_5812319 | 0.35 |
ENSDART00000114472
|
si:ch211-264f5.8
|
si:ch211-264f5.8 |
| chr1_+_18867506 | 0.35 |
ENSDART00000184180
|
rnf38
|
ring finger protein 38 |
| chr19_+_32979132 | 0.35 |
ENSDART00000169469
ENSDART00000171782 ENSDART00000180705 ENSDART00000179326 ENSDART00000193791 |
spire1a
|
spire-type actin nucleation factor 1a |
| chr5_-_38342992 | 0.35 |
ENSDART00000140337
|
mink1
|
misshapen-like kinase 1 |
| chr18_+_13248956 | 0.35 |
ENSDART00000080709
|
plcg2
|
phospholipase C, gamma 2 |
| chr13_+_11550454 | 0.35 |
ENSDART00000034935
ENSDART00000166908 |
desi2
|
desumoylating isopeptidase 2 |
| chr2_-_5014939 | 0.35 |
ENSDART00000164506
|
dlg1l
|
discs, large (Drosophila) homolog 1, like |
| chr13_+_13945218 | 0.35 |
ENSDART00000089501
ENSDART00000142997 |
eif2ak3
|
eukaryotic translation initiation factor 2-alpha kinase 3 |
| chr7_-_49801183 | 0.35 |
ENSDART00000052083
|
fjx1
|
four jointed box 1 |
| chr9_-_1639884 | 0.34 |
ENSDART00000062850
ENSDART00000150947 |
agps
|
alkylglycerone phosphate synthase |
| chr6_-_31336317 | 0.34 |
ENSDART00000193927
|
dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr9_+_38168012 | 0.34 |
ENSDART00000102445
|
clasp1a
|
cytoplasmic linker associated protein 1a |
| chr7_+_51795667 | 0.34 |
ENSDART00000174201
ENSDART00000073839 |
slc38a7
|
solute carrier family 38, member 7 |
| chr5_-_57311037 | 0.34 |
ENSDART00000149855
|
fer
|
fer (fps/fes related) tyrosine kinase |
| chr10_+_31809226 | 0.34 |
ENSDART00000087898
|
foxo1b
|
forkhead box O1 b |
| chr1_-_354115 | 0.34 |
ENSDART00000141590
ENSDART00000098627 |
pros1
|
protein S |
| chr3_-_2623176 | 0.34 |
ENSDART00000179792
ENSDART00000123512 |
si:dkey-217f16.6
|
si:dkey-217f16.6 |
| chr23_+_39346774 | 0.34 |
ENSDART00000190985
|
src
|
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| chr13_-_33194647 | 0.34 |
ENSDART00000134383
|
trip11
|
thyroid hormone receptor interactor 11 |
| chr2_-_37312927 | 0.34 |
ENSDART00000141214
|
skila
|
SKI-like proto-oncogene a |
| chr21_+_13387965 | 0.34 |
ENSDART00000134347
|
zgc:113162
|
zgc:113162 |
| chr8_+_54081819 | 0.34 |
ENSDART00000005857
ENSDART00000161795 |
prickle2a
|
prickle homolog 2a |
| chr20_-_38787341 | 0.34 |
ENSDART00000136771
|
dnajc5ga
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma a |
| chr13_-_4979029 | 0.34 |
ENSDART00000132931
|
nolc1
|
nucleolar and coiled-body phosphoprotein 1 |
| chr3_-_10634438 | 0.34 |
ENSDART00000093037
ENSDART00000130761 ENSDART00000156617 |
map2k4a
|
mitogen-activated protein kinase kinase 4a |
| chr4_+_8680767 | 0.34 |
ENSDART00000182726
|
adipor2
|
adiponectin receptor 2 |
| chr10_-_6587066 | 0.34 |
ENSDART00000171833
|
chd1
|
chromodomain helicase DNA binding protein 1 |
| chr17_+_44463230 | 0.34 |
ENSDART00000130311
|
naa30
|
N(alpha)-acetyltransferase 30, NatC catalytic subunit |
| chr6_-_24384654 | 0.34 |
ENSDART00000164723
|
brdt
|
bromodomain, testis-specific |
| chr3_+_35498119 | 0.33 |
ENSDART00000178963
|
tnrc6a
|
trinucleotide repeat containing 6a |
| chr5_-_38384289 | 0.33 |
ENSDART00000135260
|
mink1
|
misshapen-like kinase 1 |
| chr9_-_29029643 | 0.33 |
ENSDART00000151941
|
ptpn4a
|
protein tyrosine phosphatase, non-receptor type 4a |
| chr19_+_42231431 | 0.33 |
ENSDART00000102698
|
jtb
|
jumping translocation breakpoint |
| chr3_-_44753325 | 0.33 |
ENSDART00000074543
|
hs3st4
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 4 |
| chr7_-_22632518 | 0.33 |
ENSDART00000161046
|
si:dkey-112a7.4
|
si:dkey-112a7.4 |
| chr5_+_66170479 | 0.33 |
ENSDART00000172117
|
gldc
|
glycine dehydrogenase (decarboxylating) |
| chr9_-_29029980 | 0.33 |
ENSDART00000000670
|
ptpn4a
|
protein tyrosine phosphatase, non-receptor type 4a |
| chr23_+_39346930 | 0.33 |
ENSDART00000102843
|
src
|
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| chr13_-_35472531 | 0.32 |
ENSDART00000057052
|
slx4ip
|
SLX4 interacting protein |
| chr7_+_67381912 | 0.32 |
ENSDART00000167564
|
nfat5b
|
nuclear factor of activated T cells 5b |
| chr8_-_8446668 | 0.32 |
ENSDART00000132700
|
cdk16
|
cyclin-dependent kinase 16 |
| chr23_-_22523303 | 0.32 |
ENSDART00000079019
|
spsb1
|
splA/ryanodine receptor domain and SOCS box containing 1 |
| chr5_+_28260158 | 0.32 |
ENSDART00000181434
|
ncaph
|
non-SMC condensin I complex, subunit H |
| chr15_+_47746176 | 0.32 |
ENSDART00000154481
ENSDART00000160914 |
stard10
|
StAR-related lipid transfer (START) domain containing 10 |
| chr14_+_1127866 | 0.32 |
ENSDART00000172820
|
sec24b
|
SEC24 homolog B, COPII coat complex component |
| chr6_-_13022166 | 0.32 |
ENSDART00000157139
|
tmbim1a
|
transmembrane BAX inhibitor motif containing 1a |
| chr2_+_52065884 | 0.32 |
ENSDART00000146418
|
shda
|
Src homology 2 domain containing transforming protein D, a |
| chr20_-_28768109 | 0.32 |
ENSDART00000114611
ENSDART00000182443 |
sipa1l1
|
signal-induced proliferation-associated 1 like 1 |
| chr16_+_53590586 | 0.32 |
ENSDART00000083534
|
gpatch3
|
G patch domain containing 3 |
| chr9_+_34230067 | 0.32 |
ENSDART00000139157
|
nxpe3
|
neurexophilin and PC-esterase domain family, member 3 |
| chr20_+_51061695 | 0.32 |
ENSDART00000134416
|
im:7140055
|
im:7140055 |
| chr10_+_37173029 | 0.32 |
ENSDART00000136510
|
ksr1a
|
kinase suppressor of ras 1a |
| chr25_-_37191929 | 0.32 |
ENSDART00000128108
|
urahb
|
urate (5-hydroxyiso-) hydrolase b |
| chr20_-_15161502 | 0.32 |
ENSDART00000187072
|
plpp6
|
phospholipid phosphatase 6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of myf6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.3 | GO:0033206 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.3 | 1.5 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.2 | 0.7 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.2 | 0.5 | GO:1904869 | positive regulation of protein localization to nucleus(GO:1900182) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.1 | 0.6 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.1 | 0.4 | GO:0005991 | trehalose metabolic process(GO:0005991) |
| 0.1 | 2.1 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 | 0.4 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.1 | 0.4 | GO:0055109 | invagination involved in gastrulation with mouth forming second(GO:0055109) morphogenesis of an epithelial fold(GO:0060571) |
| 0.1 | 0.5 | GO:0032206 | positive regulation of telomere maintenance(GO:0032206) regulation of telomere capping(GO:1904353) positive regulation of telomere capping(GO:1904355) |
| 0.1 | 0.3 | GO:0097384 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.1 | 0.3 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.1 | 0.3 | GO:0051037 | histone displacement(GO:0001207) regulation of transcription involved in meiotic cell cycle(GO:0051037) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.1 | 0.7 | GO:0060965 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.1 | 0.4 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.1 | 0.3 | GO:0039529 | RIG-I signaling pathway(GO:0039529) regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039531) regulation of RIG-I signaling pathway(GO:0039535) |
| 0.1 | 0.3 | GO:0019284 | L-methionine biosynthetic process from S-adenosylmethionine(GO:0019284) |
| 0.1 | 0.6 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) protein urmylation(GO:0032447) |
| 0.1 | 0.3 | GO:0036135 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) Schwann cell migration(GO:0036135) cardiac neuron differentiation(GO:0060945) cardiac neuron development(GO:0060959) |
| 0.1 | 0.7 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.1 | 0.5 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.1 | 0.4 | GO:0009450 | gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.1 | 0.7 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 0.5 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.1 | 0.3 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.1 | 0.4 | GO:0010828 | positive regulation of glucose transport(GO:0010828) |
| 0.1 | 0.3 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 0.1 | 0.3 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 0.3 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.1 | 0.3 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 0.2 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 0.3 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.1 | 0.2 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.1 | 0.2 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.1 | 0.2 | GO:2000378 | negative regulation of reactive oxygen species metabolic process(GO:2000378) |
| 0.1 | 0.3 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.1 | 0.2 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.1 | 0.3 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 0.3 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.1 | 0.7 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.1 | 0.4 | GO:2000178 | negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.1 | 0.5 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.1 | 0.2 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.1 | 0.6 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 0.2 | GO:0051228 | protein hexamerization(GO:0034214) mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.1 | 0.3 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.2 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.1 | 0.2 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.1 | 0.2 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 0.8 | GO:0032204 | regulation of telomere maintenance(GO:0032204) |
| 0.1 | 0.3 | GO:0030327 | prenylated protein catabolic process(GO:0030327) prenylcysteine catabolic process(GO:0030328) prenylcysteine metabolic process(GO:0030329) |
| 0.1 | 0.3 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.1 | 0.5 | GO:0003422 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.1 | 0.4 | GO:0046959 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.1 | 0.4 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.1 | 0.4 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.1 | 0.2 | GO:0009097 | isoleucine metabolic process(GO:0006549) isoleucine biosynthetic process(GO:0009097) |
| 0.1 | 0.4 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) |
| 0.0 | 0.4 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.3 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.4 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.2 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.4 | GO:0046247 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.0 | 0.4 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.7 | GO:0035196 | production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.0 | 0.4 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.2 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.2 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 0.2 | GO:0048211 | Golgi vesicle docking(GO:0048211) |
| 0.0 | 0.2 | GO:0098920 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 0.0 | 0.2 | GO:0042148 | strand invasion(GO:0042148) |
| 0.0 | 0.5 | GO:0046620 | regulation of organ growth(GO:0046620) |
| 0.0 | 0.1 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.1 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.4 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 0.3 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.0 | 0.2 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.6 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.2 | GO:0031113 | regulation of microtubule polymerization(GO:0031113) |
| 0.0 | 0.3 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.0 | 0.2 | GO:0060368 | regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060368) |
| 0.0 | 0.3 | GO:0008207 | C21-steroid hormone metabolic process(GO:0008207) |
| 0.0 | 1.7 | GO:0048599 | oocyte development(GO:0048599) |
| 0.0 | 0.2 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.6 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.2 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.3 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.2 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.1 | GO:1903292 | protein localization to Golgi membrane(GO:1903292) |
| 0.0 | 0.2 | GO:0035513 | oxidative RNA demethylation(GO:0035513) |
| 0.0 | 0.2 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.0 | 0.2 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 | 0.3 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 1.2 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 0.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.3 | GO:2001239 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.1 | GO:0098529 | neuromuscular junction development, skeletal muscle fiber(GO:0098529) |
| 0.0 | 0.2 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.2 | GO:0045687 | positive regulation of glial cell differentiation(GO:0045687) positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.0 | 0.6 | GO:0007257 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 0.2 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 0.3 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.0 | 0.2 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.1 | GO:0003161 | cardiac conduction system development(GO:0003161) negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.4 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.2 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.4 | GO:0032785 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.3 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.2 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.4 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.0 | 0.1 | GO:0009838 | B cell apoptotic process(GO:0001783) regulation of B cell apoptotic process(GO:0002902) abscission(GO:0009838) mitotic cytokinesis checkpoint(GO:0044878) regulation of lymphocyte apoptotic process(GO:0070228) |
| 0.0 | 0.5 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.1 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.0 | 0.2 | GO:0045337 | farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.0 | 0.3 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.1 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.3 | GO:0046037 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.0 | 0.1 | GO:0060300 | regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 0.0 | 0.1 | GO:0034086 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.0 | 0.1 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.0 | 0.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.3 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 1.2 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.2 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.2 | GO:1904396 | regulation of neuromuscular junction development(GO:1904396) |
| 0.0 | 0.1 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.7 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 0.2 | GO:0045634 | regulation of melanocyte differentiation(GO:0045634) |
| 0.0 | 0.4 | GO:1903288 | positive regulation of sodium ion transport(GO:0010765) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.0 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:0019062 | virion attachment to host cell(GO:0019062) adhesion of symbiont to host(GO:0044406) adhesion of symbiont to host cell(GO:0044650) |
| 0.0 | 0.1 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.0 | 0.2 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.2 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.0 | 0.3 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.0 | 0.1 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.8 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.3 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.0 | 0.3 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.2 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.2 | GO:0071623 | negative regulation of leukocyte chemotaxis(GO:0002689) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.0 | 0.2 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.2 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.2 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.4 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.3 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.1 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.5 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.3 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.1 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.0 | 0.3 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.0 | 0.4 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.2 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.0 | 0.6 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.1 | GO:0048569 | post-embryonic organ development(GO:0048569) |
| 0.0 | 0.2 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.0 | 0.1 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.0 | 0.9 | GO:0043534 | blood vessel endothelial cell migration(GO:0043534) |
| 0.0 | 0.1 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.2 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.2 | GO:0055090 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.0 | 0.3 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.0 | 0.3 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.3 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.1 | GO:1902269 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.2 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.2 | GO:0035188 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.4 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.1 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.0 | 0.5 | GO:0099633 | protein localization to postsynaptic specialization membrane(GO:0099633) neurotransmitter receptor localization to postsynaptic specialization membrane(GO:0099645) |
| 0.0 | 0.4 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.0 | GO:0032847 | regulation of cellular pH reduction(GO:0032847) synaptic vesicle lumen acidification(GO:0097401) regulation of synaptic vesicle lumen acidification(GO:1901546) |
| 0.0 | 0.1 | GO:0016556 | mRNA modification(GO:0016556) |
| 0.0 | 0.4 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.5 | GO:2001236 | regulation of extrinsic apoptotic signaling pathway(GO:2001236) |
| 0.0 | 0.3 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.0 | 0.8 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.3 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.1 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 | 0.7 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 0.7 | GO:0042752 | regulation of circadian rhythm(GO:0042752) |
| 0.0 | 0.1 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) histone H2A K63-linked ubiquitination(GO:0070535) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.2 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.0 | 0.0 | GO:0060785 | regulation of apoptosis involved in tissue homeostasis(GO:0060785) |
| 0.0 | 0.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.1 | GO:0098787 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.5 | GO:0072114 | pronephros morphogenesis(GO:0072114) |
| 0.0 | 0.2 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.1 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.4 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.6 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.6 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 0.4 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.1 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.0 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.0 | 0.4 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.3 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.1 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.7 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 0.1 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.0 | 0.4 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 0.1 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.3 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.2 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.4 | GO:0048794 | swim bladder development(GO:0048794) |
| 0.0 | 0.2 | GO:0031647 | regulation of protein stability(GO:0031647) |
| 0.0 | 0.7 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.1 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.3 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.1 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.0 | 0.2 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.0 | GO:0036445 | neuronal stem cell division(GO:0036445) somatic stem cell division(GO:0048103) |
| 0.0 | 0.1 | GO:0009794 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.0 | 0.1 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.0 | GO:0051103 | DNA ligation(GO:0006266) DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 | 0.0 | GO:0010610 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.0 | 0.2 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.2 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.5 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.2 | 1.7 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.2 | 0.7 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.1 | 0.4 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.1 | 0.7 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.1 | 0.3 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.1 | 0.3 | GO:0031417 | NatC complex(GO:0031417) |
| 0.1 | 0.3 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.1 | 0.3 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 0.3 | GO:0031251 | PAN complex(GO:0031251) |
| 0.1 | 0.2 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.1 | 0.3 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.1 | 0.2 | GO:0005948 | acetolactate synthase complex(GO:0005948) |
| 0.1 | 0.5 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 0.5 | GO:0033061 | DNA recombinase mediator complex(GO:0033061) |
| 0.1 | 0.8 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 1.2 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.1 | 0.4 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 0.4 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.5 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.2 | GO:0008247 | 1-alkyl-2-acetylglycerophosphocholine esterase complex(GO:0008247) |
| 0.0 | 0.1 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.0 | 0.3 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.2 | GO:0046695 | SLIK (SAGA-like) complex(GO:0046695) |
| 0.0 | 0.4 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.2 | GO:1990071 | TRAPPII protein complex(GO:1990071) |
| 0.0 | 0.4 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.2 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.3 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.3 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.5 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.2 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.0 | 2.9 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 0.1 | GO:0044326 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.0 | 0.6 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.3 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.3 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.2 | GO:0000172 | ribonuclease MRP complex(GO:0000172) multimeric ribonuclease P complex(GO:0030681) |
| 0.0 | 0.2 | GO:0000938 | GARP complex(GO:0000938) |
| 0.0 | 0.5 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.2 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.6 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.1 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.2 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.6 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.1 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 1.4 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.2 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.7 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.2 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 2.1 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.1 | GO:0031213 | RSF complex(GO:0031213) |
| 0.0 | 0.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 1.4 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.1 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.0 | 0.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.0 | 1.2 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.0 | 0.2 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 0.3 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.5 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.0 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.1 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.2 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.3 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.2 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 1.6 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.0 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.0 | 0.1 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.2 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.3 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.0 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 1.8 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.4 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.2 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.1 | GO:0036156 | inner dynein arm(GO:0036156) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.2 | 1.0 | GO:0010858 | calcium-dependent protein kinase inhibitor activity(GO:0008427) calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.1 | 0.6 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.1 | 0.4 | GO:0004555 | alpha,alpha-trehalase activity(GO:0004555) trehalase activity(GO:0015927) |
| 0.1 | 0.3 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.1 | 1.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.4 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.3 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 0.4 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.1 | 0.4 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.1 | 0.5 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.1 | 0.2 | GO:0003984 | acetolactate synthase activity(GO:0003984) |
| 0.1 | 0.5 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 0.3 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.1 | 0.2 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.1 | 0.2 | GO:0004113 | 2',3'-cyclic-nucleotide 3'-phosphodiesterase activity(GO:0004113) |
| 0.1 | 2.1 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 0.6 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.1 | 0.9 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 0.2 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 0.3 | GO:0001735 | prenylcysteine oxidase activity(GO:0001735) |
| 0.1 | 0.3 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 0.2 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.1 | 0.5 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.3 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 0.3 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.1 | 0.3 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 0.1 | 0.2 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.1 | 0.2 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.0 | 0.2 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.4 | GO:0010436 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.0 | 0.2 | GO:0035516 | oxidative DNA demethylase activity(GO:0035516) |
| 0.0 | 0.2 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.3 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.1 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 0.2 | GO:0019779 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.0 | 0.2 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.0 | 0.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.7 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.3 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0017050 | D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.5 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.4 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.0 | 0.1 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.0 | 0.7 | GO:0017136 | histone deacetylase activity(GO:0004407) NAD-dependent histone deacetylase activity(GO:0017136) |
| 0.0 | 0.1 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) type II activin receptor binding(GO:0070699) |
| 0.0 | 0.1 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.2 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.3 | GO:0016594 | glycine binding(GO:0016594) |
| 0.0 | 0.4 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.2 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.2 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.0 | 0.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.0 | 0.4 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.2 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.0 | 1.9 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 0.7 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.6 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.3 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.0 | 0.2 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.0 | 0.5 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.2 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.1 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 0.4 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.1 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.0 | 1.0 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.6 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.3 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.2 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.2 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.3 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.1 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.2 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.1 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.0 | 0.4 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.3 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 1.0 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.2 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.3 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.3 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.3 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.1 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.1 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.0 | 0.1 | GO:0001047 | core promoter sequence-specific DNA binding(GO:0001046) core promoter binding(GO:0001047) |
| 0.0 | 0.6 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.4 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.3 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.4 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.2 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.4 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.4 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 1.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 2.1 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.2 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.2 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.8 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.3 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.2 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.2 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.1 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.0 | 0.2 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.3 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.1 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.2 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.2 | GO:0031995 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0008263 | mismatch base pair DNA N-glycosylase activity(GO:0000700) pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.0 | 0.2 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.4 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.4 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.3 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.4 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.3 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.4 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.4 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 1.0 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.1 | GO:1990518 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.3 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.0 | 0.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.0 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
| 0.0 | 0.0 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.2 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.0 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.8 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 0.4 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.3 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.2 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 1.2 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.4 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.9 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.8 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.1 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 1.2 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.4 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.7 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.3 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.5 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.5 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.6 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.8 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.3 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.1 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.3 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.0 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.1 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.0 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.6 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.5 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.8 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.2 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.6 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.1 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.3 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.3 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.7 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.3 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.2 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.1 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.2 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 0.1 | PID ALK2 PATHWAY | ALK2 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 0.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.5 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 0.4 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 0.3 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.5 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.1 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.7 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.3 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.4 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.3 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.1 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.3 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.2 | REACTOME SIGNALING BY ERBB4 | Genes involved in Signaling by ERBB4 |
| 0.0 | 0.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.4 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.4 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.4 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.3 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.3 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.3 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.5 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.1 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.1 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.2 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.1 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.0 | 0.3 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.1 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.3 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.1 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.2 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.4 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |