Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
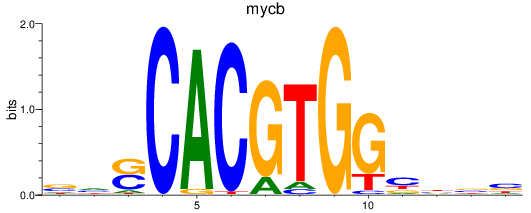
Results for mycb
Z-value: 1.21
Transcription factors associated with mycb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mycb
|
ENSDARG00000007241 | MYC proto-oncogene, bHLH transcription factor b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mycb | dr11_v1_chr2_+_32016256_32016256 | -0.31 | 4.2e-01 | Click! |
Activity profile of mycb motif
Sorted Z-values of mycb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_24555935 | 3.17 |
ENSDART00000132660
ENSDART00000162801 |
polr3gla
|
polymerase (RNA) III (DNA directed) polypeptide G like a |
| chr19_-_24555623 | 3.09 |
ENSDART00000176022
|
polr3gla
|
polymerase (RNA) III (DNA directed) polypeptide G like a |
| chr23_+_38159715 | 2.10 |
ENSDART00000137969
|
zgc:112994
|
zgc:112994 |
| chr23_-_28025943 | 1.89 |
ENSDART00000181146
|
sp5l
|
Sp5 transcription factor-like |
| chr5_-_8765428 | 1.84 |
ENSDART00000167793
|
mybbp1a
|
MYB binding protein (P160) 1a |
| chr9_-_56272465 | 1.32 |
ENSDART00000039235
|
lcp1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr5_+_68807170 | 1.25 |
ENSDART00000017849
|
her7
|
hairy and enhancer of split related-7 |
| chr1_-_19648227 | 0.96 |
ENSDART00000054574
|
polr1e
|
polymerase (RNA) I polypeptide E |
| chr9_+_38158570 | 0.94 |
ENSDART00000059549
ENSDART00000133060 |
nifk
|
nucleolar protein interacting with the FHA domain of MKI67 |
| chr14_-_12822 | 0.91 |
ENSDART00000180650
ENSDART00000188819 |
msx1a
|
muscle segment homeobox 1a |
| chr14_-_28568107 | 0.89 |
ENSDART00000042850
ENSDART00000145502 |
insb
|
preproinsulin b |
| chr22_-_26834043 | 0.87 |
ENSDART00000087202
|
si:dkey-44g23.5
|
si:dkey-44g23.5 |
| chr16_+_23960933 | 0.86 |
ENSDART00000146077
|
apoeb
|
apolipoprotein Eb |
| chr2_+_18988407 | 0.84 |
ENSDART00000170216
|
glula
|
glutamate-ammonia ligase (glutamine synthase) a |
| chr16_+_23960744 | 0.79 |
ENSDART00000058965
|
apoeb
|
apolipoprotein Eb |
| chr9_-_12652984 | 0.76 |
ENSDART00000052256
|
sumo3b
|
small ubiquitin-like modifier 3b |
| chr8_+_46386601 | 0.75 |
ENSDART00000129661
ENSDART00000084081 |
ogg1
|
8-oxoguanine DNA glycosylase |
| chr11_+_3959495 | 0.74 |
ENSDART00000122953
|
gnl3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr13_-_45022301 | 0.74 |
ENSDART00000183589
ENSDART00000125633 ENSDART00000074787 |
khdrbs1a
|
KH domain containing, RNA binding, signal transduction associated 1a |
| chr16_+_23961276 | 0.69 |
ENSDART00000192754
|
apoeb
|
apolipoprotein Eb |
| chr13_-_9367647 | 0.66 |
ENSDART00000083362
ENSDART00000144146 |
si:dkey-33c12.4
|
si:dkey-33c12.4 |
| chr4_-_890220 | 0.66 |
ENSDART00000022668
|
aim1b
|
crystallin beta-gamma domain containing 1b |
| chr5_-_61609448 | 0.65 |
ENSDART00000133426
|
si:dkey-261j4.5
|
si:dkey-261j4.5 |
| chr11_+_6456146 | 0.65 |
ENSDART00000036939
|
gadd45ba
|
growth arrest and DNA-damage-inducible, beta a |
| chr19_+_42328423 | 0.65 |
ENSDART00000077059
|
anp32e
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr17_+_24318753 | 0.64 |
ENSDART00000064083
|
otx1
|
orthodenticle homeobox 1 |
| chr21_-_30408775 | 0.62 |
ENSDART00000101037
|
nhp2
|
NHP2 ribonucleoprotein homolog (yeast) |
| chr10_+_39283985 | 0.62 |
ENSDART00000016464
|
dcps
|
decapping enzyme, scavenger |
| chr17_-_48705993 | 0.61 |
ENSDART00000030934
|
kcnk5a
|
potassium channel, subfamily K, member 5a |
| chr13_-_4992395 | 0.57 |
ENSDART00000102651
|
nolc1
|
nucleolar and coiled-body phosphoprotein 1 |
| chr10_+_29849977 | 0.55 |
ENSDART00000180242
|
hspa8
|
heat shock protein 8 |
| chr19_-_791016 | 0.54 |
ENSDART00000037515
|
msto1
|
misato 1, mitochondrial distribution and morphology regulator |
| chr10_-_39283883 | 0.52 |
ENSDART00000023831
|
cry5
|
cryptochrome circadian clock 5 |
| chr23_+_25135858 | 0.51 |
ENSDART00000103986
|
fam3a
|
family with sequence similarity 3, member A |
| chr2_-_42375275 | 0.51 |
ENSDART00000026339
|
gtpbp4
|
GTP binding protein 4 |
| chr22_-_607812 | 0.50 |
ENSDART00000145983
|
cdkn1a
|
cyclin-dependent kinase inhibitor 1A |
| chr15_+_6652396 | 0.49 |
ENSDART00000192813
ENSDART00000157678 |
nop53
|
NOP53 ribosome biogenesis factor |
| chr13_+_31402067 | 0.49 |
ENSDART00000019202
|
tdrd9
|
tudor domain containing 9 |
| chr24_+_36317544 | 0.49 |
ENSDART00000048640
ENSDART00000156096 |
pus3
|
pseudouridylate synthase 3 |
| chr12_-_7607114 | 0.47 |
ENSDART00000158095
|
slc16a9b
|
solute carrier family 16, member 9b |
| chr9_-_11587070 | 0.47 |
ENSDART00000030995
|
umps
|
uridine monophosphate synthetase |
| chr19_+_1005933 | 0.46 |
ENSDART00000191953
|
zdhhc3b
|
zinc finger, DHHC-type containing 3b |
| chr8_-_2246143 | 0.45 |
ENSDART00000135835
|
si:dkeyp-117b11.3
|
si:dkeyp-117b11.3 |
| chr9_+_29985010 | 0.44 |
ENSDART00000020743
|
cmss1
|
cms1 ribosomal small subunit homolog (yeast) |
| chr5_+_3927989 | 0.44 |
ENSDART00000030125
|
znhit3
|
zinc finger, HIT-type containing 3 |
| chr4_-_2945306 | 0.44 |
ENSDART00000128934
ENSDART00000019518 |
aebp2
|
AE binding protein 2 |
| chr1_+_17376922 | 0.43 |
ENSDART00000145068
|
fat1a
|
FAT atypical cadherin 1a |
| chr10_+_29850330 | 0.43 |
ENSDART00000168898
|
hspa8
|
heat shock protein 8 |
| chr12_-_4301234 | 0.43 |
ENSDART00000152377
ENSDART00000152521 |
ca15b
|
carbonic anhydrase XVb |
| chr25_+_17405458 | 0.43 |
ENSDART00000186711
|
e2f4
|
E2F transcription factor 4 |
| chr3_+_24595922 | 0.42 |
ENSDART00000169405
|
si:dkey-68o6.5
|
si:dkey-68o6.5 |
| chr20_+_25625872 | 0.42 |
ENSDART00000078385
|
ppat
|
phosphoribosyl pyrophosphate amidotransferase |
| chr9_-_11676491 | 0.41 |
ENSDART00000022358
|
zc3h15
|
zinc finger CCCH-type containing 15 |
| chr15_+_42235449 | 0.41 |
ENSDART00000114801
ENSDART00000182053 |
SGPP2
|
sphingosine-1-phosphate phosphatase 2 |
| chr25_+_17405201 | 0.41 |
ENSDART00000164349
|
e2f4
|
E2F transcription factor 4 |
| chr8_-_46386024 | 0.40 |
ENSDART00000136602
ENSDART00000060919 ENSDART00000137472 |
qars
|
glutaminyl-tRNA synthetase |
| chr1_-_54972170 | 0.40 |
ENSDART00000150548
ENSDART00000038330 |
khsrp
|
KH-type splicing regulatory protein |
| chr17_+_13099476 | 0.40 |
ENSDART00000012670
|
pnn
|
pinin, desmosome associated protein |
| chr8_-_23599096 | 0.39 |
ENSDART00000183096
|
slc38a5b
|
solute carrier family 38, member 5b |
| chr19_-_26964348 | 0.39 |
ENSDART00000103955
|
zbtb12.2
|
zinc finger and BTB domain containing 12, tandem duplicate 2 |
| chr11_+_25296366 | 0.38 |
ENSDART00000065949
|
top1l
|
DNA topoisomerase I, like |
| chr11_-_30636163 | 0.38 |
ENSDART00000140516
|
zgc:153665
|
zgc:153665 |
| chr6_-_10752937 | 0.38 |
ENSDART00000135093
|
ola1
|
Obg-like ATPase 1 |
| chr20_-_25631256 | 0.38 |
ENSDART00000048164
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr9_-_27391908 | 0.37 |
ENSDART00000135221
|
nepro
|
nucleolus and neural progenitor protein |
| chr4_+_90048 | 0.37 |
ENSDART00000166440
|
lrp6
|
low density lipoprotein receptor-related protein 6 |
| chr6_-_39080630 | 0.37 |
ENSDART00000021520
ENSDART00000128308 |
eif4bb
|
eukaryotic translation initiation factor 4Bb |
| chr24_-_39518599 | 0.36 |
ENSDART00000145606
ENSDART00000031486 |
lyrm1
|
LYR motif containing 1 |
| chr14_+_22076596 | 0.36 |
ENSDART00000106147
ENSDART00000100278 ENSDART00000131489 |
slc43a1a
|
solute carrier family 43 (amino acid system L transporter), member 1a |
| chr21_-_14832369 | 0.36 |
ENSDART00000144859
|
pus1
|
pseudouridylate synthase 1 |
| chr20_+_25445826 | 0.36 |
ENSDART00000012581
|
pfas
|
phosphoribosylformylglycinamidine synthase |
| chr1_-_54971968 | 0.35 |
ENSDART00000140016
|
khsrp
|
KH-type splicing regulatory protein |
| chr20_-_25645150 | 0.34 |
ENSDART00000063137
|
si:dkeyp-117h8.4
|
si:dkeyp-117h8.4 |
| chr23_-_38160024 | 0.34 |
ENSDART00000087112
|
pfdn4
|
prefoldin subunit 4 |
| chr17_+_51682429 | 0.33 |
ENSDART00000004379
|
nol10
|
nucleolar protein 10 |
| chr25_+_30442389 | 0.33 |
ENSDART00000003346
|
pdcd2l
|
programmed cell death 2-like |
| chr14_+_16287968 | 0.32 |
ENSDART00000106593
|
prpf19
|
pre-mRNA processing factor 19 |
| chr23_-_21535040 | 0.32 |
ENSDART00000010647
|
rcc2
|
regulator of chromosome condensation 2 |
| chr4_+_36489448 | 0.32 |
ENSDART00000143181
|
znf1149
|
zinc finger protein 1149 |
| chr5_-_3927692 | 0.32 |
ENSDART00000146840
ENSDART00000058346 |
c1qbp
|
complement component 1, q subcomponent binding protein |
| chr6_-_58975010 | 0.32 |
ENSDART00000144911
ENSDART00000144514 |
mars
|
methionyl-tRNA synthetase |
| chr3_-_23575007 | 0.31 |
ENSDART00000155282
ENSDART00000087726 |
igf2bp1
|
insulin-like growth factor 2 mRNA binding protein 1 |
| chr22_+_6293563 | 0.31 |
ENSDART00000063416
|
rnasel2
|
ribonuclease like 2 |
| chr11_+_25485774 | 0.31 |
ENSDART00000026249
|
gnl3l
|
guanine nucleotide binding protein-like 3 (nucleolar)-like |
| chr9_-_21460164 | 0.30 |
ENSDART00000133469
|
zmym2
|
zinc finger, MYM-type 2 |
| chr23_-_21534455 | 0.30 |
ENSDART00000139092
|
rcc2
|
regulator of chromosome condensation 2 |
| chr21_-_45382112 | 0.30 |
ENSDART00000151029
ENSDART00000151335 ENSDART00000151687 ENSDART00000075438 |
cdkn2aipnl
|
CDKN2A interacting protein N-terminal like |
| chr16_-_31790285 | 0.29 |
ENSDART00000184655
|
chd4b
|
chromodomain helicase DNA binding protein 4b |
| chr18_-_5850683 | 0.29 |
ENSDART00000082087
|
nip7
|
NIP7, nucleolar pre-rRNA processing protein |
| chr5_-_54714789 | 0.28 |
ENSDART00000063357
|
ccnb1
|
cyclin B1 |
| chr7_+_1550966 | 0.28 |
ENSDART00000177863
ENSDART00000126840 |
supt16h
|
SPT16 homolog, facilitates chromatin remodeling subunit |
| chr14_+_52408619 | 0.28 |
ENSDART00000163856
|
noa1
|
nitric oxide associated 1 |
| chr22_+_5118361 | 0.26 |
ENSDART00000168371
ENSDART00000170222 ENSDART00000158846 |
mibp
|
muscle-specific beta 1 integrin binding protein |
| chr25_-_8513255 | 0.26 |
ENSDART00000150129
|
polg
|
polymerase (DNA directed), gamma |
| chr23_-_21534738 | 0.26 |
ENSDART00000134587
|
rcc2
|
regulator of chromosome condensation 2 |
| chr1_-_14533611 | 0.25 |
ENSDART00000180678
|
CR847936.6
|
|
| chr13_+_13930263 | 0.25 |
ENSDART00000079154
|
rpia
|
ribose 5-phosphate isomerase A (ribose 5-phosphate epimerase) |
| chr19_-_22346582 | 0.25 |
ENSDART00000045675
ENSDART00000169065 |
slc52a2
zgc:109744
|
solute carrier family 52 (riboflavin transporter), member 2 zgc:109744 |
| chr24_+_32668675 | 0.25 |
ENSDART00000156638
ENSDART00000155973 |
si:ch211-282b22.1
|
si:ch211-282b22.1 |
| chr22_+_10713713 | 0.25 |
ENSDART00000122349
|
hiat1b
|
hippocampus abundant transcript 1b |
| chr1_-_35916247 | 0.24 |
ENSDART00000181541
|
smad1
|
SMAD family member 1 |
| chr21_+_3941758 | 0.24 |
ENSDART00000181345
|
setx
|
senataxin |
| chr19_-_32804535 | 0.23 |
ENSDART00000175613
ENSDART00000052098 |
nt5c1aa
|
5'-nucleotidase, cytosolic IAa |
| chr16_-_21668082 | 0.23 |
ENSDART00000088513
|
gnl1
|
guanine nucleotide binding protein-like 1 |
| chr8_-_52859301 | 0.23 |
ENSDART00000162004
|
nr5a1a
|
nuclear receptor subfamily 5, group A, member 1a |
| chr25_+_245438 | 0.22 |
ENSDART00000004689
|
zgc:92481
|
zgc:92481 |
| chr16_+_16968682 | 0.22 |
ENSDART00000111074
|
si:ch211-120k19.1
|
si:ch211-120k19.1 |
| chr1_+_34203817 | 0.22 |
ENSDART00000191432
ENSDART00000046094 |
arl6
|
ADP-ribosylation factor-like 6 |
| chr5_+_72194444 | 0.22 |
ENSDART00000165436
|
ddx54
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 54 |
| chr18_+_14277003 | 0.22 |
ENSDART00000006628
|
zgc:173742
|
zgc:173742 |
| chr2_-_10014821 | 0.22 |
ENSDART00000185525
|
CR956623.1
|
|
| chr7_-_26462831 | 0.22 |
ENSDART00000113543
|
mblac1
|
metallo-beta-lactamase domain containing 1 |
| chr6_-_54078623 | 0.21 |
ENSDART00000154076
|
hyal1
|
hyaluronoglucosaminidase 1 |
| chr8_-_40251126 | 0.21 |
ENSDART00000180435
|
kdm2ba
|
lysine (K)-specific demethylase 2Ba |
| chr15_-_44052927 | 0.21 |
ENSDART00000166209
|
wu:fb44b02
|
wu:fb44b02 |
| chr4_+_18824959 | 0.21 |
ENSDART00000146141
ENSDART00000040424 |
slc26a3.1
|
solute carrier family 26 (anion exchanger), member 3 |
| chr4_-_14192254 | 0.21 |
ENSDART00000143804
|
pus7l
|
pseudouridylate synthase 7-like |
| chr14_+_20941038 | 0.20 |
ENSDART00000182539
|
zgc:66433
|
zgc:66433 |
| chr7_+_41322407 | 0.20 |
ENSDART00000114076
ENSDART00000139093 |
dph2
|
DPH2 homolog (S. cerevisiae) |
| chr2_+_19236969 | 0.19 |
ENSDART00000163875
ENSDART00000168644 |
cc2d1b
|
coiled-coil and C2 domain containing 1B |
| chr16_+_16969060 | 0.19 |
ENSDART00000182819
ENSDART00000191876 |
si:ch211-120k19.1
rpl18
|
si:ch211-120k19.1 ribosomal protein L18 |
| chr6_-_39653972 | 0.18 |
ENSDART00000155859
|
larp4ab
|
La ribonucleoprotein domain family, member 4Ab |
| chr11_-_12800945 | 0.18 |
ENSDART00000191178
|
txlng
|
taxilin gamma |
| chr21_-_39577517 | 0.18 |
ENSDART00000143566
|
srsf1b
|
serine/arginine-rich splicing factor 1b |
| chr9_+_33788389 | 0.18 |
ENSDART00000144623
|
kdm6a
|
lysine (K)-specific demethylase 6A |
| chr11_-_12801157 | 0.17 |
ENSDART00000103449
|
txlng
|
taxilin gamma |
| chr19_+_791538 | 0.17 |
ENSDART00000146554
ENSDART00000138406 |
tmem79a
|
transmembrane protein 79a |
| chr25_+_19095231 | 0.17 |
ENSDART00000154066
|
isg20
|
interferon stimulated exonuclease gene |
| chr17_-_45370200 | 0.17 |
ENSDART00000186208
|
znf106a
|
zinc finger protein 106a |
| chr8_+_23355484 | 0.16 |
ENSDART00000085361
ENSDART00000125729 |
dnmt3ba
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate a |
| chr1_-_6494384 | 0.16 |
ENSDART00000109356
|
klf7a
|
Kruppel-like factor 7a |
| chr24_-_42090635 | 0.16 |
ENSDART00000166413
|
ssr1
|
signal sequence receptor, alpha |
| chr4_+_61171310 | 0.16 |
ENSDART00000141738
|
si:dkey-9p20.18
|
si:dkey-9p20.18 |
| chr6_-_13114406 | 0.16 |
ENSDART00000188015
|
zgc:194469
|
zgc:194469 |
| chr12_-_3453589 | 0.16 |
ENSDART00000175918
|
CABZ01063170.1
|
|
| chr4_-_38033800 | 0.15 |
ENSDART00000159662
|
si:dkeyp-82b4.4
|
si:dkeyp-82b4.4 |
| chr9_-_27719998 | 0.15 |
ENSDART00000161068
ENSDART00000148195 ENSDART00000138386 |
gtf2e1
|
general transcription factor IIE, polypeptide 1, alpha |
| chr9_-_27720612 | 0.15 |
ENSDART00000000566
|
gtf2e1
|
general transcription factor IIE, polypeptide 1, alpha |
| chr5_-_54714525 | 0.15 |
ENSDART00000150138
ENSDART00000150070 |
ccnb1
|
cyclin B1 |
| chr13_-_48431766 | 0.15 |
ENSDART00000159688
ENSDART00000171765 |
fbxo11a
|
F-box protein 11a |
| chr21_+_38638979 | 0.14 |
ENSDART00000143373
|
rbmx2
|
RNA binding motif protein, X-linked 2 |
| chr25_+_37443194 | 0.14 |
ENSDART00000163178
ENSDART00000190262 |
slc10a3
|
solute carrier family 10, member 3 |
| chr3_-_40955780 | 0.14 |
ENSDART00000130130
|
cyp3c3
|
cytochrome P450, family 3, subfamily c, polypeptide 3 |
| chr8_-_13419049 | 0.14 |
ENSDART00000133656
|
pimr101
|
Pim proto-oncogene, serine/threonine kinase, related 101 |
| chr20_-_52882881 | 0.13 |
ENSDART00000111078
|
wu:fi04e12
|
wu:fi04e12 |
| chr19_+_7757682 | 0.13 |
ENSDART00000092138
ENSDART00000092112 |
ubap2l
|
ubiquitin associated protein 2-like |
| chr8_-_13471916 | 0.13 |
ENSDART00000146558
|
pimr105
|
Pim proto-oncogene, serine/threonine kinase, related 105 |
| chr13_-_18345854 | 0.13 |
ENSDART00000080107
|
si:dkey-228d14.5
|
si:dkey-228d14.5 |
| chr2_+_9946121 | 0.12 |
ENSDART00000100696
|
tsen15
|
TSEN15 tRNA splicing endonuclease subunit |
| chr21_-_11199366 | 0.12 |
ENSDART00000167666
|
dnajc21
|
DnaJ (Hsp40) homolog, subfamily C, member 21 |
| chr5_-_41933912 | 0.12 |
ENSDART00000097574
|
ncor1
|
nuclear receptor corepressor 1 |
| chr10_+_16036573 | 0.12 |
ENSDART00000188757
|
lmnb1
|
lamin B1 |
| chr4_-_44611273 | 0.12 |
ENSDART00000156793
|
znf1115
|
zinc finger protein 1115 |
| chr22_+_28969071 | 0.11 |
ENSDART00000163427
|
pimr95
|
Pim proto-oncogene, serine/threonine kinase, related 95 |
| chr21_-_34032650 | 0.11 |
ENSDART00000138575
ENSDART00000047515 |
rnf145b
|
ring finger protein 145b |
| chr9_+_27720428 | 0.11 |
ENSDART00000112415
|
lcmt2
|
leucine carboxyl methyltransferase 2 |
| chr15_-_43978141 | 0.10 |
ENSDART00000041249
|
chordc1a
|
cysteine and histidine-rich domain (CHORD) containing 1a |
| chr10_+_16036246 | 0.10 |
ENSDART00000141586
ENSDART00000135868 ENSDART00000065037 ENSDART00000124502 |
lmnb1
|
lamin B1 |
| chr13_+_15702142 | 0.10 |
ENSDART00000135960
|
trmt61a
|
tRNA methyltransferase 61A |
| chr9_-_904227 | 0.10 |
ENSDART00000144068
|
zgc:101851
|
zgc:101851 |
| chr19_+_24882845 | 0.10 |
ENSDART00000010580
|
si:ch211-195b13.1
|
si:ch211-195b13.1 |
| chr13_+_48359573 | 0.09 |
ENSDART00000161959
ENSDART00000165311 |
msh6
|
mutS homolog 6 (E. coli) |
| chr13_-_35892051 | 0.09 |
ENSDART00000145884
|
tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr1_-_26675969 | 0.09 |
ENSDART00000054184
|
trmo
|
tRNA methyltransferase O |
| chr16_-_38629208 | 0.09 |
ENSDART00000126705
|
eif3ea
|
eukaryotic translation initiation factor 3, subunit E, a |
| chr4_-_64709908 | 0.08 |
ENSDART00000161032
|
si:dkey-9i5.2
|
si:dkey-9i5.2 |
| chr10_-_14929630 | 0.08 |
ENSDART00000121892
ENSDART00000044756 ENSDART00000128579 ENSDART00000147653 |
smad2
|
SMAD family member 2 |
| chr2_+_19236677 | 0.08 |
ENSDART00000166292
|
cc2d1b
|
coiled-coil and C2 domain containing 1B |
| chr10_+_428269 | 0.08 |
ENSDART00000140715
|
zdhhc8a
|
zinc finger, DHHC-type containing 8a |
| chr1_-_26676391 | 0.08 |
ENSDART00000152492
|
trmo
|
tRNA methyltransferase O |
| chr21_+_45387903 | 0.08 |
ENSDART00000186253
ENSDART00000075432 |
jade2
|
jade family PHD finger 2 |
| chr16_+_13822137 | 0.07 |
ENSDART00000163251
|
flcn
|
folliculin |
| chr1_+_31674297 | 0.07 |
ENSDART00000044214
|
wbp1lb
|
WW domain binding protein 1-like b |
| chr14_+_4796168 | 0.07 |
ENSDART00000041468
|
ap1ar
|
adaptor-related protein complex 1 associated regulatory protein |
| chr13_+_39277178 | 0.07 |
ENSDART00000113259
|
si:dkey-85a20.4
|
si:dkey-85a20.4 |
| chr13_-_3516473 | 0.06 |
ENSDART00000146240
|
prkn
|
parkin RBR E3 ubiquitin protein ligase |
| chr24_+_39518774 | 0.06 |
ENSDART00000132939
|
dcun1d3
|
defective in cullin neddylation 1 domain containing 3 |
| chr22_+_17261801 | 0.06 |
ENSDART00000192978
ENSDART00000193187 ENSDART00000179953 ENSDART00000134798 |
tdrd5
|
tudor domain containing 5 |
| chr14_+_5385855 | 0.06 |
ENSDART00000031508
|
lbx2
|
ladybird homeobox 2 |
| chr2_-_56387041 | 0.06 |
ENSDART00000036240
|
cers4b
|
ceramide synthase 4b |
| chr5_-_1999417 | 0.06 |
ENSDART00000155437
ENSDART00000145781 |
si:ch211-160e1.5
|
si:ch211-160e1.5 |
| chr8_-_30979494 | 0.06 |
ENSDART00000138959
|
si:ch211-251j10.3
|
si:ch211-251j10.3 |
| chr21_+_45510448 | 0.05 |
ENSDART00000160494
ENSDART00000167914 |
fnip1
|
folliculin interacting protein 1 |
| chr9_+_1339194 | 0.05 |
ENSDART00000186976
ENSDART00000014766 |
casp8l2
|
caspase 8, apoptosis-related cysteine peptidase, like 2 |
| chr24_+_16905188 | 0.05 |
ENSDART00000066760
|
cct5
|
chaperonin containing TCP1, subunit 5 (epsilon) |
| chr15_-_2632891 | 0.04 |
ENSDART00000081840
|
cldnj
|
claudin j |
| chr9_-_12888082 | 0.04 |
ENSDART00000133135
ENSDART00000134415 |
si:ch211-167j6.3
|
si:ch211-167j6.3 |
| chr20_-_48470599 | 0.04 |
ENSDART00000166857
|
CABZ01059120.1
|
|
| chr21_+_19517492 | 0.04 |
ENSDART00000123168
ENSDART00000187993 |
gzmk
|
granzyme K |
| chr22_+_1440702 | 0.04 |
ENSDART00000165677
|
si:dkeyp-53d3.3
|
si:dkeyp-53d3.3 |
| chr7_+_33136545 | 0.04 |
ENSDART00000173485
|
itln2
|
intelectin 2 |
| chr18_+_20225961 | 0.04 |
ENSDART00000045679
|
tle3a
|
transducin-like enhancer of split 3a |
| chr1_-_29061285 | 0.03 |
ENSDART00000053933
ENSDART00000142350 ENSDART00000192615 |
gemin8
|
gem (nuclear organelle) associated protein 8 |
| chr9_+_907459 | 0.03 |
ENSDART00000034850
ENSDART00000144114 |
dbi
|
diazepam binding inhibitor (GABA receptor modulator, acyl-CoA binding protein) |
| chr13_-_280652 | 0.03 |
ENSDART00000193627
|
slc30a6
|
solute carrier family 30 (zinc transporter), member 6 |
| chr7_-_28463106 | 0.02 |
ENSDART00000137799
|
trim66
|
tripartite motif containing 66 |
| chr5_-_16475374 | 0.02 |
ENSDART00000134274
ENSDART00000136004 |
piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr15_+_44053244 | 0.02 |
ENSDART00000059550
|
lrrc51
|
leucine rich repeat containing 51 |
| chr3_-_47038481 | 0.02 |
ENSDART00000193260
|
CR392045.1
|
|
| chr3_+_57991074 | 0.02 |
ENSDART00000076077
|
myadml2
|
myeloid-associated differentiation marker-like 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of mycb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0071831 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 0.2 | 0.8 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.2 | 0.6 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.2 | 6.3 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.2 | 1.0 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.2 | 1.0 | GO:1902946 | positive regulation of receptor-mediated endocytosis(GO:0048260) protein localization to early endosome(GO:1902946) |
| 0.1 | 0.8 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 1.0 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 | 0.3 | GO:0001120 | DNA-templated transcriptional open complex formation(GO:0001112) transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) protein-DNA complex remodeling(GO:0001120) |
| 0.1 | 0.4 | GO:1902024 | L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.1 | 0.5 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.1 | 0.7 | GO:1904035 | epithelial cell apoptotic process(GO:1904019) regulation of epithelial cell apoptotic process(GO:1904035) |
| 0.1 | 0.6 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 0.5 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 0.3 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.1 | 0.3 | GO:0070131 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
| 0.1 | 1.4 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 0.7 | GO:0000467 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.1 | 0.5 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.1 | 0.7 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.1 | 1.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 1.9 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.1 | 0.4 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.1 | 0.3 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.4 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 1.3 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.3 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.3 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.0 | 0.5 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.0 | 0.6 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.1 | GO:0031591 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.0 | 0.6 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.2 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.2 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.1 | GO:0016446 | somatic diversification of immune receptors via somatic mutation(GO:0002566) pyrimidine dimer repair(GO:0006290) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.3 | GO:0034724 | DNA replication-independent nucleosome organization(GO:0034724) |
| 0.0 | 0.6 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.1 | GO:1904105 | positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.0 | 0.3 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.0 | 0.9 | GO:0043049 | otic placode formation(GO:0043049) |
| 0.0 | 0.6 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.8 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.5 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.6 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.2 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.0 | 0.5 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.4 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.2 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.1 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) |
| 0.0 | 0.3 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.0 | 0.2 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.3 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.4 | GO:0044332 | Wnt signaling pathway involved in dorsal/ventral axis specification(GO:0044332) |
| 0.0 | 0.1 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.1 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.5 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.2 | GO:0009651 | response to salt stress(GO:0009651) |
| 0.0 | 0.3 | GO:0042255 | ribosome assembly(GO:0042255) |
| 0.0 | 0.2 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.1 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.3 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.1 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.3 | GO:0015804 | neutral amino acid transport(GO:0015804) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.3 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.3 | 6.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.2 | 1.0 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.2 | 0.6 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.1 | 0.4 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.1 | 0.6 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 0.3 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.1 | 0.3 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 0.3 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.3 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.3 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.1 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.0 | 1.3 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 8.5 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.3 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.1 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.0 | 0.1 | GO:0071540 | eukaryotic translation initiation factor 3 complex, eIF3e(GO:0071540) |
| 0.0 | 0.5 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.1 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.1 | GO:0016580 | Sin3 complex(GO:0016580) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.2 | 0.6 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.2 | 0.7 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.2 | 0.5 | GO:0003913 | DNA photolyase activity(GO:0003913) |
| 0.1 | 0.8 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 0.5 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 0.4 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.1 | 0.2 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.1 | 0.6 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.1 | 0.8 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 1.0 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.4 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 0.5 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 1.0 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.4 | GO:0033592 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.0 | 0.8 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.6 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.3 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.3 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) |
| 0.0 | 0.3 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.0 | 1.0 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.3 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.7 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.4 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.1 | GO:0032404 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.0 | 0.4 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.6 | GO:0022842 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.3 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.2 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.2 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.3 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.6 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.5 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.5 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.5 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.4 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.1 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.4 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.6 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.1 | 1.0 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 1.0 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 0.7 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 0.6 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.4 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.5 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.9 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.7 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.5 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.6 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.1 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.4 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.9 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.3 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |