Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
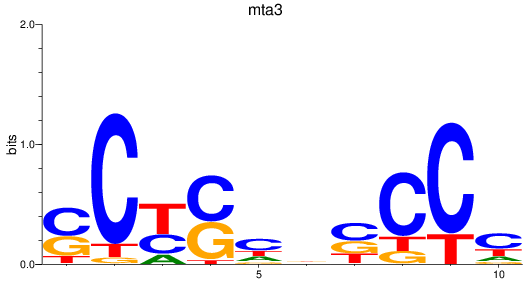
Results for mta3
Z-value: 0.83
Transcription factors associated with mta3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mta3
|
ENSDARG00000054903 | metastasis associated 1 family, member 3 |
|
mta3
|
ENSDARG00000113436 | metastasis associated 1 family, member 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mta3 | dr11_v1_chr12_+_25223843_25223873 | 0.85 | 3.9e-03 | Click! |
Activity profile of mta3 motif
Sorted Z-values of mta3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_2578026 | 1.95 |
ENSDART00000065821
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr5_-_54712159 | 1.32 |
ENSDART00000149207
|
ccnb1
|
cyclin B1 |
| chr1_+_15062 | 1.12 |
ENSDART00000169005
|
cep97
|
centrosomal protein 97 |
| chr5_+_45139196 | 1.11 |
ENSDART00000113738
|
smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr5_-_20678300 | 0.94 |
ENSDART00000088639
|
wscd2
|
WSC domain containing 2 |
| chr1_+_14454663 | 0.92 |
ENSDART00000005067
|
rbpja
|
recombination signal binding protein for immunoglobulin kappa J region a |
| chr23_+_2825940 | 0.90 |
ENSDART00000135781
|
plcg1
|
phospholipase C, gamma 1 |
| chr5_+_45138934 | 0.90 |
ENSDART00000041412
ENSDART00000136002 |
smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr5_-_23800376 | 0.90 |
ENSDART00000134184
|
gbgt1l4
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 4 |
| chr21_+_44581654 | 0.89 |
ENSDART00000187201
ENSDART00000180360 ENSDART00000191543 ENSDART00000180039 ENSDART00000186308 |
TMEM164
|
transmembrane protein 164 |
| chr22_+_26703026 | 0.89 |
ENSDART00000158756
|
crebbpa
|
CREB binding protein a |
| chr15_-_25198107 | 0.83 |
ENSDART00000159793
|
mnta
|
MAX network transcriptional repressor a |
| chr20_-_33675676 | 0.82 |
ENSDART00000147168
|
rock2b
|
rho-associated, coiled-coil containing protein kinase 2b |
| chr12_+_13348918 | 0.80 |
ENSDART00000181373
|
rnasen
|
ribonuclease type III, nuclear |
| chr5_+_20453874 | 0.77 |
ENSDART00000124545
ENSDART00000008402 |
sart3
|
squamous cell carcinoma antigen recognized by T cells 3 |
| chr22_-_193234 | 0.77 |
ENSDART00000131067
|
fbxo42
|
F-box protein 42 |
| chr20_+_9474841 | 0.76 |
ENSDART00000053847
|
rad51b
|
RAD51 paralog B |
| chr17_-_25630822 | 0.75 |
ENSDART00000126201
ENSDART00000105503 ENSDART00000151878 |
rab3gap2
|
RAB3 GTPase activating protein subunit 2 (non-catalytic) |
| chr6_+_3334392 | 0.74 |
ENSDART00000133707
ENSDART00000130879 |
st3gal3a
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3a |
| chr7_+_1579236 | 0.71 |
ENSDART00000172830
|
supt16h
|
SPT16 homolog, facilitates chromatin remodeling subunit |
| chr4_-_13255700 | 0.70 |
ENSDART00000162277
ENSDART00000026593 |
grip1
|
glutamate receptor interacting protein 1 |
| chr25_+_27744293 | 0.69 |
ENSDART00000103519
ENSDART00000149456 |
wasla
|
Wiskott-Aldrich syndrome-like a |
| chr13_+_23093743 | 0.67 |
ENSDART00000148034
|
pik3ap1
|
phosphoinositide-3-kinase adaptor protein 1 |
| chr12_-_290413 | 0.59 |
ENSDART00000152496
|
adprm
|
ADP-ribose/CDP-alcohol diphosphatase, manganese-dependent |
| chr17_-_8312923 | 0.57 |
ENSDART00000064678
|
lft2
|
lefty2 |
| chr3_+_16566361 | 0.55 |
ENSDART00000132869
ENSDART00000080771 |
slc6a16a
|
solute carrier family 6, member 16a |
| chr6_-_12270226 | 0.53 |
ENSDART00000180473
|
pkp4
|
plakophilin 4 |
| chr13_+_31545812 | 0.53 |
ENSDART00000076527
|
ppm1aa
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Aa |
| chr14_-_237130 | 0.52 |
ENSDART00000164988
|
bod1l1
|
biorientation of chromosomes in cell division 1-like 1 |
| chr10_-_9115383 | 0.50 |
ENSDART00000139324
|
si:dkeyp-41f9.3
|
si:dkeyp-41f9.3 |
| chr6_+_3334710 | 0.42 |
ENSDART00000132848
|
st3gal3a
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3a |
| chr5_+_66433287 | 0.42 |
ENSDART00000170757
|
kntc1
|
kinetochore associated 1 |
| chr16_-_280835 | 0.40 |
ENSDART00000190541
|
LO017917.1
|
|
| chr13_-_10431476 | 0.35 |
ENSDART00000133968
|
camkmt
|
calmodulin-lysine N-methyltransferase |
| chr4_+_69577362 | 0.34 |
ENSDART00000160913
|
znf1030
|
zinc finger protein 1030 |
| chr7_+_1467863 | 0.30 |
ENSDART00000173433
|
emc4
|
ER membrane protein complex subunit 4 |
| chr25_-_35143360 | 0.27 |
ENSDART00000188033
|
zgc:165555
|
zgc:165555 |
| chr20_-_54435287 | 0.26 |
ENSDART00000148632
|
yy1b
|
YY1 transcription factor b |
| chr20_-_51727860 | 0.23 |
ENSDART00000147044
|
brox
|
BRO1 domain and CAAX motif containing |
| chr2_-_16359042 | 0.19 |
ENSDART00000057216
|
arhgef4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr13_+_1100197 | 0.18 |
ENSDART00000139560
|
ppp3r1a
|
protein phosphatase 3, regulatory subunit B, alpha a |
| chr22_-_367569 | 0.14 |
ENSDART00000041895
|
ssu72
|
SSU72 homolog, RNA polymerase II CTD phosphatase |
| chr3_-_5228137 | 0.09 |
ENSDART00000137105
|
myh9b
|
myosin, heavy chain 9b, non-muscle |
| chr7_+_17560808 | 0.07 |
ENSDART00000159699
ENSDART00000165724 ENSDART00000129017 ENSDART00000101955 |
nitr1b
|
novel immune-type receptor 1b |
| chr11_-_20956309 | 0.06 |
ENSDART00000188659
|
CABZ01008739.1
|
|
| chr20_-_14665002 | 0.06 |
ENSDART00000152816
|
scrn2
|
secernin 2 |
| chr5_-_38170996 | 0.05 |
ENSDART00000145805
|
si:ch211-284e13.12
|
si:ch211-284e13.12 |
| chr25_-_35106751 | 0.04 |
ENSDART00000180682
ENSDART00000073457 |
zgc:194989
|
zgc:194989 |
| chr21_+_45712247 | 0.04 |
ENSDART00000163982
|
camlg
|
calcium modulating ligand |
| chr23_-_24825863 | 0.02 |
ENSDART00000112493
|
syt6a
|
synaptotagmin VIa |
| chr14_-_2369849 | 0.01 |
ENSDART00000180422
ENSDART00000189731 ENSDART00000111748 |
pcdhb
|
protocadherin b |
Network of associatons between targets according to the STRING database.
First level regulatory network of mta3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.0 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.3 | 0.9 | GO:0035477 | regulation of angioblast cell migration involved in selective angioblast sprouting(GO:0035477) |
| 0.3 | 0.8 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.2 | 0.9 | GO:0072149 | glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.1 | 1.3 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.1 | 1.9 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.5 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 | 0.7 | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.1 | 0.9 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.7 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.3 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.0 | 0.6 | GO:0034724 | DNA replication-independent nucleosome organization(GO:0034724) |
| 0.0 | 0.8 | GO:1901888 | regulation of cell junction assembly(GO:1901888) |
| 0.0 | 0.6 | GO:0003128 | heart field specification(GO:0003128) |
| 0.0 | 0.5 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.5 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.3 | GO:0055090 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.0 | 0.1 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.2 | 0.8 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.1 | 0.6 | GO:0035101 | FACT complex(GO:0035101) |
| 0.1 | 2.0 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.8 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.5 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.3 | GO:0072546 | ER membrane protein complex(GO:0072546) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.9 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.2 | 1.2 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.1 | 0.8 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.1 | 0.8 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 0.8 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.1 | 0.7 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 0.7 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.6 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.6 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.5 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 2.0 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 0.3 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.0 | 0.9 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 1.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.9 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.5 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.6 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.1 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 3.3 | PID CMYB PATHWAY | C-MYB transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 1.3 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.6 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.7 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |