Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
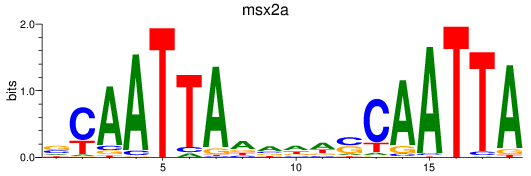
Results for msx2a
Z-value: 0.77
Transcription factors associated with msx2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
msx2a
|
ENSDARG00000104651 | muscle segment homeobox 2a |
|
msx2a
|
ENSDARG00000115813 | muscle segment homeobox 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| msx2a | dr11_v1_chr14_-_24081929_24081929 | 0.78 | 1.4e-02 | Click! |
Activity profile of msx2a motif
Sorted Z-values of msx2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_7666021 | 2.41 |
ENSDART00000180007
|
CABZ01021592.1
|
|
| chr16_-_55028740 | 2.22 |
ENSDART00000156368
ENSDART00000161704 |
zgc:114181
|
zgc:114181 |
| chr14_+_46313396 | 1.74 |
ENSDART00000047525
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr12_-_25916530 | 1.20 |
ENSDART00000186386
|
sncgb
|
synuclein, gamma b (breast cancer-specific protein 1) |
| chr9_+_53337974 | 1.06 |
ENSDART00000145138
|
dct
|
dopachrome tautomerase |
| chr10_-_29900546 | 1.04 |
ENSDART00000147441
|
lim2.1
|
lens intrinsic membrane protein 2.1 |
| chr25_+_20089986 | 1.03 |
ENSDART00000143441
ENSDART00000184073 |
tnni4b.2
|
troponin I4b, tandem duplicate 2 |
| chr15_+_28685625 | 1.02 |
ENSDART00000188797
ENSDART00000166036 |
nova2
|
neuro-oncological ventral antigen 2 |
| chr1_-_38756870 | 1.00 |
ENSDART00000130324
ENSDART00000148404 |
gpm6ab
|
glycoprotein M6Ab |
| chr13_-_6081803 | 0.99 |
ENSDART00000099224
|
dld
|
deltaD |
| chr1_+_52929185 | 0.98 |
ENSDART00000147683
|
inpp4b
|
inositol polyphosphate-4-phosphatase type II B |
| chr1_+_10051763 | 0.94 |
ENSDART00000011701
|
fgb
|
fibrinogen beta chain |
| chr5_-_63515210 | 0.88 |
ENSDART00000022348
|
prdm12b
|
PR domain containing 12b |
| chr1_+_53374454 | 0.85 |
ENSDART00000038807
|
ucp1
|
uncoupling protein 1 |
| chr6_+_52804267 | 0.84 |
ENSDART00000065681
|
matn4
|
matrilin 4 |
| chr4_+_2252123 | 0.84 |
ENSDART00000163996
ENSDART00000066491 |
glipr1a
|
GLI pathogenesis-related 1a |
| chr14_-_498979 | 0.82 |
ENSDART00000171976
|
spry1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
| chr25_+_14507567 | 0.80 |
ENSDART00000015681
|
dbx1b
|
developing brain homeobox 1b |
| chr5_+_11840905 | 0.77 |
ENSDART00000030444
|
tesca
|
tescalcin a |
| chr12_+_47663419 | 0.76 |
ENSDART00000171932
|
HHEX
|
hematopoietically expressed homeobox |
| chr6_+_7123980 | 0.76 |
ENSDART00000179738
ENSDART00000151311 |
si:ch211-237c6.4
|
si:ch211-237c6.4 |
| chr3_-_30941362 | 0.74 |
ENSDART00000076830
|
coro1a
|
coronin, actin binding protein, 1A |
| chr4_-_1324141 | 0.73 |
ENSDART00000180720
|
ptn
|
pleiotrophin |
| chr24_+_17269849 | 0.71 |
ENSDART00000017605
|
spag6
|
sperm associated antigen 6 |
| chr16_+_10918252 | 0.70 |
ENSDART00000172949
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr5_+_56268436 | 0.67 |
ENSDART00000021159
|
lhx1b
|
LIM homeobox 1b |
| chr2_+_54641644 | 0.67 |
ENSDART00000027313
|
ndufv2
|
NADH dehydrogenase (ubiquinone) flavoprotein 2 |
| chr4_+_14660769 | 0.67 |
ENSDART00000168152
ENSDART00000013990 ENSDART00000079987 |
abcc9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr11_+_3989403 | 0.67 |
ENSDART00000082379
|
spcs1
|
signal peptidase complex subunit 1 |
| chr21_+_5129513 | 0.66 |
ENSDART00000102572
|
thbs4b
|
thrombospondin 4b |
| chr20_+_38032143 | 0.65 |
ENSDART00000032161
|
galnt14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr21_-_16113477 | 0.65 |
ENSDART00000147588
|
cyb561a3b
|
cytochrome b561 family, member A3b |
| chr1_-_35924495 | 0.64 |
ENSDART00000184424
|
smad1
|
SMAD family member 1 |
| chr9_-_32753535 | 0.63 |
ENSDART00000060006
|
olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr1_-_22678471 | 0.63 |
ENSDART00000128918
|
fgfbp1b
|
fibroblast growth factor binding protein 1b |
| chr6_+_25257728 | 0.62 |
ENSDART00000162581
|
kyat3
|
kynurenine aminotransferase 3 |
| chr24_+_17270129 | 0.61 |
ENSDART00000186729
|
spag6
|
sperm associated antigen 6 |
| chr16_-_13921589 | 0.59 |
ENSDART00000023543
|
rcvrn2
|
recoverin 2 |
| chr7_-_26408472 | 0.58 |
ENSDART00000111494
|
gal3st4
|
galactose-3-O-sulfotransferase 4 |
| chr17_+_41992054 | 0.58 |
ENSDART00000182878
ENSDART00000111537 |
kiz
|
kizuna centrosomal protein |
| chr20_-_53321499 | 0.58 |
ENSDART00000179894
ENSDART00000127427 ENSDART00000084952 |
wasf1
|
WAS protein family, member 1 |
| chr23_+_19813677 | 0.57 |
ENSDART00000139192
ENSDART00000142308 |
emd
|
emerin (Emery-Dreifuss muscular dystrophy) |
| chr21_+_29077509 | 0.56 |
ENSDART00000128561
|
ecscr
|
endothelial cell surface expressed chemotaxis and apoptosis regulator |
| chr7_+_15872357 | 0.55 |
ENSDART00000165757
|
pax6b
|
paired box 6b |
| chr16_-_26255877 | 0.54 |
ENSDART00000146214
|
erfl1
|
Ets2 repressor factor like 1 |
| chr24_-_7699356 | 0.54 |
ENSDART00000013117
|
syt5b
|
synaptotagmin Vb |
| chr6_-_39270851 | 0.54 |
ENSDART00000148839
|
arhgef25b
|
Rho guanine nucleotide exchange factor (GEF) 25b |
| chr16_-_28878080 | 0.52 |
ENSDART00000149501
|
si:dkey-239n17.4
|
si:dkey-239n17.4 |
| chr8_-_31062811 | 0.51 |
ENSDART00000142528
|
slc20a1a
|
solute carrier family 20, member 1a |
| chr2_-_10192459 | 0.50 |
ENSDART00000128535
ENSDART00000017173 |
dmbx1a
|
diencephalon/mesencephalon homeobox 1a |
| chr21_+_5589923 | 0.50 |
ENSDART00000160885
|
stbd1
|
starch binding domain 1 |
| chr13_-_37109987 | 0.50 |
ENSDART00000136750
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr8_+_40644838 | 0.50 |
ENSDART00000169311
|
adra2b
|
adrenoceptor alpha 2B |
| chr20_+_38031439 | 0.50 |
ENSDART00000153208
|
galnt14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr20_+_22220988 | 0.46 |
ENSDART00000049204
|
kdr
|
kinase insert domain receptor (a type III receptor tyrosine kinase) |
| chr10_+_20128267 | 0.44 |
ENSDART00000064615
|
dmtn
|
dematin actin binding protein |
| chr24_+_14713776 | 0.43 |
ENSDART00000134475
|
gdap1
|
ganglioside induced differentiation associated protein 1 |
| chr16_+_32238020 | 0.42 |
ENSDART00000017562
|
klhl32
|
kelch-like family member 32 |
| chr3_-_50954607 | 0.42 |
ENSDART00000163810
|
cdc42ep4a
|
CDC42 effector protein (Rho GTPase binding) 4a |
| chr17_+_43032529 | 0.42 |
ENSDART00000055611
ENSDART00000154863 |
isca2
|
iron-sulfur cluster assembly 2 |
| chr10_-_1788376 | 0.41 |
ENSDART00000123842
|
epb41l4a
|
erythrocyte membrane protein band 4.1 like 4A |
| chr3_-_58226583 | 0.41 |
ENSDART00000187429
|
im:6904045
|
im:6904045 |
| chr18_-_35837673 | 0.40 |
ENSDART00000059360
|
hnrnpul1
|
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr17_+_24613255 | 0.39 |
ENSDART00000064738
|
atp5if1b
|
ATP synthase inhibitory factor subunit 1b |
| chr14_-_4076480 | 0.39 |
ENSDART00000059231
|
enpp6
|
ectonucleotide pyrophosphatase/phosphodiesterase 6 |
| chr5_+_36752943 | 0.38 |
ENSDART00000017138
|
exoc3l2a
|
exocyst complex component 3-like 2a |
| chr20_-_9436521 | 0.37 |
ENSDART00000133000
|
zgc:101840
|
zgc:101840 |
| chr5_-_26834511 | 0.37 |
ENSDART00000136713
ENSDART00000192932 ENSDART00000113246 |
si:ch211-102c2.4
|
si:ch211-102c2.4 |
| chr14_-_1998501 | 0.36 |
ENSDART00000189052
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr8_-_2230128 | 0.35 |
ENSDART00000140427
|
si:dkeyp-117b11.2
|
si:dkeyp-117b11.2 |
| chr4_-_23908802 | 0.34 |
ENSDART00000138873
|
celf2
|
cugbp, Elav-like family member 2 |
| chr22_-_31060579 | 0.34 |
ENSDART00000182376
|
cand2
|
cullin-associated and neddylation-dissociated 2 (putative) |
| chr3_+_27798094 | 0.34 |
ENSDART00000075100
ENSDART00000151437 |
carhsp1
|
calcium regulated heat stable protein 1 |
| chr4_-_9557186 | 0.34 |
ENSDART00000150569
|
shank3b
|
SH3 and multiple ankyrin repeat domains 3b |
| chr3_-_35541378 | 0.34 |
ENSDART00000183075
ENSDART00000022147 |
ndufab1b
|
NADH dehydrogenase (ubiquinone) 1, alpha/beta subcomplex, 1b |
| chr20_-_4738101 | 0.33 |
ENSDART00000050201
ENSDART00000152559 ENSDART00000053858 ENSDART00000125620 |
papola
|
poly(A) polymerase alpha |
| chr24_-_38787457 | 0.33 |
ENSDART00000112318
ENSDART00000154461 |
ntn2
|
netrin 2 |
| chr2_+_42212948 | 0.32 |
ENSDART00000147019
|
si:ch211-235o23.1
|
si:ch211-235o23.1 |
| chr7_+_69653981 | 0.32 |
ENSDART00000090165
|
frem1a
|
Fras1 related extracellular matrix 1a |
| chr10_-_20524592 | 0.32 |
ENSDART00000185048
|
ddhd2
|
DDHD domain containing 2 |
| chr18_-_17485419 | 0.32 |
ENSDART00000018764
|
foxl1
|
forkhead box L1 |
| chr4_+_5796761 | 0.32 |
ENSDART00000164854
|
si:ch73-352p4.8
|
si:ch73-352p4.8 |
| chr15_-_34214440 | 0.32 |
ENSDART00000167052
|
etv1
|
ets variant 1 |
| chr14_+_36869273 | 0.31 |
ENSDART00000111674
|
F2RL3
|
si:ch211-132p1.2 |
| chr15_-_28094256 | 0.31 |
ENSDART00000142041
ENSDART00000132153 ENSDART00000146657 ENSDART00000048720 |
cryba1a
|
crystallin, beta A1a |
| chr7_+_34688527 | 0.30 |
ENSDART00000108473
|
plekhg4
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 4 |
| chr15_-_22074315 | 0.30 |
ENSDART00000149830
|
drd2a
|
dopamine receptor D2a |
| chr25_+_37126921 | 0.30 |
ENSDART00000124331
|
si:ch1073-174d20.1
|
si:ch1073-174d20.1 |
| chr8_-_34762163 | 0.30 |
ENSDART00000114080
|
setd1bb
|
SET domain containing 1B, b |
| chr17_+_21452258 | 0.29 |
ENSDART00000157098
|
pla2g4f.1
|
phospholipase A2, group IVF, tandem duplicate 1 |
| chr2_-_49985351 | 0.28 |
ENSDART00000145888
|
ccr12a
|
chemokine (C-C motif) receptor 12a |
| chr7_+_59169081 | 0.28 |
ENSDART00000167980
|
ostc
|
oligosaccharyltransferase complex subunit |
| chr5_+_2815021 | 0.28 |
ENSDART00000020472
|
hpda
|
4-hydroxyphenylpyruvate dioxygenase a |
| chr23_+_16935494 | 0.27 |
ENSDART00000143120
|
si:dkey-147f3.4
|
si:dkey-147f3.4 |
| chr24_+_17007407 | 0.27 |
ENSDART00000110652
|
zfx
|
zinc finger protein, X-linked |
| chr13_+_1369003 | 0.27 |
ENSDART00000146082
|
ifngr1
|
interferon gamma receptor 1 |
| chr24_+_11381400 | 0.26 |
ENSDART00000058703
|
ackr4b
|
atypical chemokine receptor 4b |
| chr20_-_38836161 | 0.26 |
ENSDART00000061358
|
si:dkey-221h15.4
|
si:dkey-221h15.4 |
| chr7_-_49351706 | 0.26 |
ENSDART00000174151
|
brsk2b
|
BR serine/threonine kinase 2b |
| chr25_+_35019693 | 0.25 |
ENSDART00000046218
|
flnca
|
filamin C, gamma a (actin binding protein 280) |
| chr2_-_1569250 | 0.25 |
ENSDART00000167202
|
dab1b
|
Dab, reelin signal transducer, homolog 1b (Drosophila) |
| chr25_+_7784582 | 0.25 |
ENSDART00000155016
|
dgkzb
|
diacylglycerol kinase, zeta b |
| chr25_+_7229046 | 0.25 |
ENSDART00000149965
ENSDART00000041820 |
lingo1a
|
leucine rich repeat and Ig domain containing 1a |
| chr14_+_597532 | 0.24 |
ENSDART00000159805
|
LO018568.2
|
|
| chr2_+_36701322 | 0.24 |
ENSDART00000002510
|
golim4b
|
golgi integral membrane protein 4b |
| chr6_+_55174744 | 0.23 |
ENSDART00000023562
|
SYT2
|
synaptotagmin 2 |
| chr4_-_25271455 | 0.22 |
ENSDART00000066936
|
tmem110l
|
transmembrane protein 110, like |
| chr14_-_1987121 | 0.22 |
ENSDART00000135733
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr10_+_40762200 | 0.22 |
ENSDART00000161000
ENSDART00000136185 |
taar19g
|
trace amine associated receptor 19g |
| chr7_+_34687969 | 0.22 |
ENSDART00000182013
ENSDART00000188255 ENSDART00000182728 |
plekhg4
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 4 |
| chr10_-_20524387 | 0.21 |
ENSDART00000159060
|
ddhd2
|
DDHD domain containing 2 |
| chr21_-_39639954 | 0.21 |
ENSDART00000026766
|
aldocb
|
aldolase C, fructose-bisphosphate, b |
| chr5_+_12528693 | 0.21 |
ENSDART00000051670
|
rfc5
|
replication factor C (activator 1) 5 |
| chr20_+_9828769 | 0.21 |
ENSDART00000160480
ENSDART00000053844 ENSDART00000080691 |
stxbp6l
|
syntaxin binding protein 6 (amisyn), like |
| chr25_+_19095231 | 0.20 |
ENSDART00000154066
|
isg20
|
interferon stimulated exonuclease gene |
| chr1_-_17569793 | 0.20 |
ENSDART00000125125
|
acsl1a
|
acyl-CoA synthetase long chain family member 1a |
| chr11_-_2632747 | 0.19 |
ENSDART00000008133
|
si:ch211-160o17.2
|
si:ch211-160o17.2 |
| chr1_+_8111009 | 0.19 |
ENSDART00000152192
|
SLC5A10
|
si:dkeyp-9d4.5 |
| chr15_-_576135 | 0.19 |
ENSDART00000124170
|
cbln20
|
cerebellin 20 |
| chr18_+_13837746 | 0.19 |
ENSDART00000169552
|
cdh13
|
cadherin 13, H-cadherin (heart) |
| chr20_-_27711970 | 0.19 |
ENSDART00000139637
|
zbtb25
|
zinc finger and BTB domain containing 25 |
| chr1_-_17570013 | 0.18 |
ENSDART00000146946
|
acsl1a
|
acyl-CoA synthetase long chain family member 1a |
| chr23_-_39784368 | 0.18 |
ENSDART00000110282
|
si:ch211-286f9.2
|
si:ch211-286f9.2 |
| chr8_+_10561922 | 0.18 |
ENSDART00000133348
|
fam19a5l
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5-like |
| chr22_+_37631234 | 0.18 |
ENSDART00000007346
|
psmd1
|
proteasome 26S subunit, non-ATPase 1 |
| chr11_-_29563437 | 0.18 |
ENSDART00000163958
|
arhgef10la
|
Rho guanine nucleotide exchange factor (GEF) 10-like a |
| chr10_+_40606084 | 0.17 |
ENSDART00000133119
|
si:ch211-238p8.24
|
si:ch211-238p8.24 |
| chr23_-_1660708 | 0.17 |
ENSDART00000175138
|
CU693481.1
|
|
| chr5_+_22177033 | 0.16 |
ENSDART00000131223
|
fdx1b
|
ferredoxin 1b |
| chr19_-_31584444 | 0.16 |
ENSDART00000052183
|
zgc:111986
|
zgc:111986 |
| chr15_+_8767650 | 0.16 |
ENSDART00000033871
|
ap2s1
|
adaptor-related protein complex 2, sigma 1 subunit |
| chr10_-_42751641 | 0.16 |
ENSDART00000182734
ENSDART00000113926 |
zgc:100918
|
zgc:100918 |
| chr17_+_35362851 | 0.15 |
ENSDART00000137659
|
cmpk2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial |
| chr10_+_40700311 | 0.15 |
ENSDART00000157650
ENSDART00000138342 |
taar19n
|
trace amine associated receptor 19n |
| chr6_+_34870374 | 0.15 |
ENSDART00000149356
|
il23r
|
interleukin 23 receptor |
| chr22_-_28668442 | 0.15 |
ENSDART00000182377
|
col8a1b
|
collagen, type VIII, alpha 1b |
| chr13_-_17723417 | 0.15 |
ENSDART00000183834
|
vdac2
|
voltage-dependent anion channel 2 |
| chr13_+_40686133 | 0.15 |
ENSDART00000146112
|
hps1
|
Hermansky-Pudlak syndrome 1 |
| chr4_+_2267641 | 0.15 |
ENSDART00000165503
|
si:ch73-89b15.3
|
si:ch73-89b15.3 |
| chr10_+_40629616 | 0.14 |
ENSDART00000147476
|
CR396590.9
|
|
| chr19_+_43604256 | 0.14 |
ENSDART00000151080
ENSDART00000110305 |
si:ch211-199g17.9
|
si:ch211-199g17.9 |
| chr21_-_35419486 | 0.14 |
ENSDART00000138529
|
si:dkeyp-23e4.3
|
si:dkeyp-23e4.3 |
| chr11_-_165288 | 0.14 |
ENSDART00000108703
ENSDART00000173151 |
tegt
|
testis enhanced gene transcript (BAX inhibitor 1) |
| chr10_+_40633990 | 0.14 |
ENSDART00000190489
ENSDART00000139474 |
si:ch211-238p8.31
|
si:ch211-238p8.31 |
| chr9_-_55790458 | 0.13 |
ENSDART00000161247
|
sowahca
|
sosondowah ankyrin repeat domain family Ca |
| chr7_+_34790768 | 0.13 |
ENSDART00000075116
|
si:dkey-148a17.6
|
si:dkey-148a17.6 |
| chr8_-_38616712 | 0.13 |
ENSDART00000141827
|
si:ch211-198d23.1
|
si:ch211-198d23.1 |
| chr20_-_35040041 | 0.13 |
ENSDART00000131919
|
kif26bb
|
kinesin family member 26Bb |
| chr19_+_43604643 | 0.12 |
ENSDART00000151168
|
si:ch211-199g17.9
|
si:ch211-199g17.9 |
| chr21_-_21089781 | 0.12 |
ENSDART00000144361
|
ank1b
|
ankyrin 1, erythrocytic b |
| chr9_+_28598577 | 0.12 |
ENSDART00000142623
ENSDART00000135947 |
si:ch73-7i4.1
|
si:ch73-7i4.1 |
| chr22_+_26853254 | 0.12 |
ENSDART00000182487
|
tmem186
|
transmembrane protein 186 |
| chr10_-_5847904 | 0.11 |
ENSDART00000161096
|
ankrd55
|
ankyrin repeat domain 55 |
| chr10_+_40756352 | 0.11 |
ENSDART00000156210
ENSDART00000144576 |
taar19f
|
trace amine associated receptor 19f |
| chr1_+_58067815 | 0.11 |
ENSDART00000156678
|
si:ch211-114l13.4
|
si:ch211-114l13.4 |
| chr19_-_10612774 | 0.11 |
ENSDART00000091898
|
si:dkey-211g8.6
|
si:dkey-211g8.6 |
| chr20_+_26556372 | 0.11 |
ENSDART00000187179
ENSDART00000157291 |
irf4b
|
interferon regulatory factor 4b |
| chr10_-_5847655 | 0.10 |
ENSDART00000192773
|
ankrd55
|
ankyrin repeat domain 55 |
| chr4_+_77907740 | 0.10 |
ENSDART00000172216
|
si:zfos-2131b9.2
|
si:zfos-2131b9.2 |
| chr2_-_54039293 | 0.09 |
ENSDART00000166013
|
abhd8a
|
abhydrolase domain containing 8a |
| chr20_+_45893173 | 0.09 |
ENSDART00000131169
|
bmp2b
|
bone morphogenetic protein 2b |
| chr10_-_34870667 | 0.09 |
ENSDART00000161272
|
dclk1a
|
doublecortin-like kinase 1a |
| chr14_+_45675306 | 0.09 |
ENSDART00000105461
|
rom1b
|
retinal outer segment membrane protein 1b |
| chr12_-_19007834 | 0.08 |
ENSDART00000153248
|
chadlb
|
chondroadherin-like b |
| chr6_-_7123210 | 0.08 |
ENSDART00000041304
|
atg3
|
autophagy related 3 |
| chr16_+_51326237 | 0.08 |
ENSDART00000168525
|
CU469384.1
|
|
| chr7_-_7493758 | 0.07 |
ENSDART00000036703
|
pfdn2
|
prefoldin subunit 2 |
| chr10_-_40448736 | 0.07 |
ENSDART00000137644
ENSDART00000168190 |
taar20p
|
trace amine associated receptor 20p |
| chr8_-_16788626 | 0.07 |
ENSDART00000191652
|
CR759968.2
|
|
| chr10_-_40508527 | 0.07 |
ENSDART00000121514
ENSDART00000133729 |
taar20b1
|
trace amine associated receptor 20b1 |
| chr17_-_11329959 | 0.07 |
ENSDART00000015418
|
irf2bpl
|
interferon regulatory factor 2 binding protein-like |
| chr4_+_19700308 | 0.06 |
ENSDART00000027919
|
pax4
|
paired box 4 |
| chr3_+_27929505 | 0.06 |
ENSDART00000055528
|
TMEM114
|
transmembrane protein 114 |
| chr10_-_40474628 | 0.06 |
ENSDART00000133892
|
taar20a1
|
trace amine associated receptor 20a1 |
| chr7_+_4474880 | 0.06 |
ENSDART00000143528
|
si:dkey-83f18.14
|
si:dkey-83f18.14 |
| chr22_+_30447969 | 0.06 |
ENSDART00000193066
|
si:dkey-169i5.4
|
si:dkey-169i5.4 |
| chr10_+_40742685 | 0.05 |
ENSDART00000184858
ENSDART00000140300 |
taar19e
|
trace amine associated receptor 19e |
| chr19_-_24224142 | 0.05 |
ENSDART00000136409
ENSDART00000114390 |
prf1.8
|
perforin 1.8 |
| chr3_+_28502419 | 0.05 |
ENSDART00000151081
|
sept12
|
septin 12 |
| chr15_-_6966221 | 0.05 |
ENSDART00000165487
ENSDART00000027657 |
mrps22
|
mitochondrial ribosomal protein S22 |
| chr21_-_40250500 | 0.05 |
ENSDART00000141008
|
si:ch211-218m3.9
|
si:ch211-218m3.9 |
| chr10_+_40774215 | 0.05 |
ENSDART00000131493
|
taar19b
|
trace amine associated receptor 19b |
| chr14_-_22385779 | 0.05 |
ENSDART00000192211
ENSDART00000175309 |
fbxl3l
|
F-box and leucine-rich repeat protein 3, like |
| chr4_-_6459863 | 0.05 |
ENSDART00000138367
|
foxp2
|
forkhead box P2 |
| chr1_-_17735861 | 0.04 |
ENSDART00000018238
|
pdlim3a
|
PDZ and LIM domain 3a |
| chr10_-_40528229 | 0.04 |
ENSDART00000146927
ENSDART00000148354 |
taar18g
|
trace amine associated receptor 18g |
| chr10_+_40726399 | 0.03 |
ENSDART00000142812
|
taar19j
|
trace amine associated receptor 19j |
| chr20_-_35557902 | 0.03 |
ENSDART00000134753
|
adgrf7
|
adhesion G protein-coupled receptor F7 |
| chr10_+_40583930 | 0.03 |
ENSDART00000134523
|
taar18j
|
trace amine associated receptor 18j |
| chr10_+_40690411 | 0.03 |
ENSDART00000146865
|
taar19m
|
trace amine associated receptor 19m |
| chr15_-_41486793 | 0.03 |
ENSDART00000138525
|
si:ch211-187g4.1
|
si:ch211-187g4.1 |
| chr15_-_29348212 | 0.02 |
ENSDART00000133117
|
tsku
|
tsukushi small leucine rich proteoglycan homolog (Xenopus laevis) |
| chr10_-_40443722 | 0.02 |
ENSDART00000132906
ENSDART00000157598 |
taar20q
|
trace amine associated receptor 20q |
| chr20_-_32188897 | 0.02 |
ENSDART00000133887
|
si:ch211-51a19.5
|
si:ch211-51a19.5 |
| chr17_-_36841981 | 0.02 |
ENSDART00000131566
|
myo6b
|
myosin VIb |
| chr5_+_38752287 | 0.01 |
ENSDART00000133571
|
cxcl11.8
|
chemokine (C-X-C motif) ligand 11, duplicate 8 |
| chr20_-_26822522 | 0.01 |
ENSDART00000146326
ENSDART00000046764 ENSDART00000103234 ENSDART00000143267 |
gmds
|
GDP-mannose 4,6-dehydratase |
| chr24_+_9881219 | 0.01 |
ENSDART00000036204
|
cdv3
|
carnitine deficiency-associated gene expressed in ventricle 3 |
| chr7_-_66868543 | 0.01 |
ENSDART00000149680
|
ampd3a
|
adenosine monophosphate deaminase 3a |
| chr25_-_18739924 | 0.01 |
ENSDART00000156328
|
si:dkeyp-93a5.2
|
si:dkeyp-93a5.2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of msx2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:2000648 | neuroblast division(GO:0055057) positive regulation of stem cell proliferation(GO:2000648) |
| 0.3 | 0.9 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.3 | 0.8 | GO:0003156 | regulation of organ formation(GO:0003156) |
| 0.2 | 0.6 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.1 | 1.0 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.1 | 0.4 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.1 | 0.4 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.1 | 0.3 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.1 | 0.7 | GO:0035989 | tendon development(GO:0035989) |
| 0.1 | 0.8 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.1 | 0.5 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.1 | 0.6 | GO:0090104 | pancreatic D cell differentiation(GO:0003311) pancreatic epsilon cell differentiation(GO:0090104) |
| 0.1 | 0.4 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 0.9 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.1 | 0.6 | GO:0035479 | angioblast cell migration from lateral mesoderm to midline(GO:0035479) |
| 0.1 | 0.6 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.1 | 0.2 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.1 | 1.2 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.3 | GO:0098815 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.0 | 0.3 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 1.0 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.4 | GO:0031272 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.7 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.2 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.1 | GO:0090008 | hypoblast development(GO:0090008) |
| 0.0 | 0.4 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.7 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.3 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.2 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.5 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.0 | 0.2 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.0 | 0.2 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.0 | 0.2 | GO:0034650 | glucocorticoid biosynthetic process(GO:0006704) cortisol metabolic process(GO:0034650) |
| 0.0 | 0.3 | GO:0051967 | negative regulation of cytosolic calcium ion concentration(GO:0051481) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.5 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 1.0 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.2 | GO:0046959 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.0 | 0.2 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 0.4 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 2.0 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.4 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.3 | GO:0070193 | synaptonemal complex assembly(GO:0007130) synaptonemal complex organization(GO:0070193) |
| 0.0 | 0.3 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.0 | 0.5 | GO:0035924 | cellular response to vascular endothelial growth factor stimulus(GO:0035924) |
| 0.0 | 0.8 | GO:0021515 | cell differentiation in spinal cord(GO:0021515) |
| 0.0 | 0.2 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.1 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.3 | GO:0098508 | endothelial to hematopoietic transition(GO:0098508) |
| 0.0 | 0.0 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.3 | GO:0043631 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.0 | 0.7 | GO:0051781 | positive regulation of cell division(GO:0051781) |
| 0.0 | 0.3 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.3 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.2 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.2 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.9 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.2 | 0.7 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 1.1 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 1.0 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 0.2 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.0 | 0.5 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.1 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.0 | 0.6 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 1.2 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.2 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 1.0 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 1.0 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.6 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.3 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.3 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.3 | 0.9 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.2 | 2.2 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.2 | 0.8 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.2 | 1.1 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 1.0 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.1 | 0.7 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.1 | 0.3 | GO:0000035 | acyl binding(GO:0000035) |
| 0.1 | 0.5 | GO:2001070 | starch binding(GO:2001070) |
| 0.1 | 0.3 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.1 | 0.5 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.1 | 0.3 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 0.4 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 0.6 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 0.2 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.3 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 1.0 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 3.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.5 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 1.3 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.4 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.5 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.6 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.2 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.1 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.0 | 0.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.1 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.5 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.3 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.2 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.5 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.3 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.5 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 0.9 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 0.3 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.6 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.7 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.6 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.0 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 0.9 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 0.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 0.7 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.5 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.3 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 1.0 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 1.2 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.7 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 1.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 0.3 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.2 | REACTOME BOTULINUM NEUROTOXICITY | Genes involved in Botulinum neurotoxicity |
| 0.0 | 0.3 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.2 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |