Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
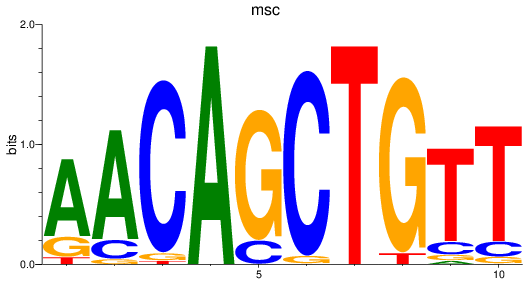
Results for msc
Z-value: 1.40
Transcription factors associated with msc
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
msc
|
ENSDARG00000110016 | musculin (activated B-cell factor-1) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| msc | dr11_v1_chr24_+_13735616_13735616 | 0.63 | 6.8e-02 | Click! |
Activity profile of msc motif
Sorted Z-values of msc motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_44715224 | 3.53 |
ENSDART00000184630
|
si:dkey-56m19.5
|
si:dkey-56m19.5 |
| chr11_-_5865744 | 3.34 |
ENSDART00000104360
|
gamt
|
guanidinoacetate N-methyltransferase |
| chr5_-_19400166 | 3.15 |
ENSDART00000008994
|
foxn4
|
forkhead box N4 |
| chr21_+_27382893 | 2.95 |
ENSDART00000005682
|
actn3a
|
actinin alpha 3a |
| chr5_-_40734045 | 2.65 |
ENSDART00000010896
|
isl1
|
ISL LIM homeobox 1 |
| chr7_-_24364536 | 2.62 |
ENSDART00000064789
|
txn
|
thioredoxin |
| chr1_+_52560549 | 2.43 |
ENSDART00000167514
|
abca1a
|
ATP-binding cassette, sub-family A (ABC1), member 1A |
| chr17_-_14836320 | 2.41 |
ENSDART00000157051
|
nid2a
|
nidogen 2a (osteonidogen) |
| chr5_-_38451082 | 2.39 |
ENSDART00000136428
|
chrne
|
cholinergic receptor, nicotinic, epsilon |
| chr13_+_42124566 | 2.38 |
ENSDART00000074707
|
cdc42ep3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr6_+_29217392 | 2.37 |
ENSDART00000006386
|
atp1b1a
|
ATPase Na+/K+ transporting subunit beta 1a |
| chr8_-_44298964 | 2.37 |
ENSDART00000098520
|
fzd10
|
frizzled class receptor 10 |
| chr4_-_4834347 | 2.24 |
ENSDART00000141803
|
coa6
|
cytochrome c oxidase assembly factor 6 |
| chr15_+_33989181 | 2.21 |
ENSDART00000169487
|
vwde
|
von Willebrand factor D and EGF domains |
| chr6_+_2097690 | 2.18 |
ENSDART00000193770
|
tgm2b
|
transglutaminase 2b |
| chr10_-_22803740 | 2.16 |
ENSDART00000079469
ENSDART00000187968 ENSDART00000122543 |
pcolcea
|
procollagen C-endopeptidase enhancer a |
| chr20_-_48485354 | 2.15 |
ENSDART00000124040
ENSDART00000148437 |
insm1a
|
insulinoma-associated 1a |
| chr11_-_41966854 | 2.14 |
ENSDART00000055709
|
her2
|
hairy-related 2 |
| chr7_+_6969909 | 2.13 |
ENSDART00000189886
|
actn3b
|
actinin alpha 3b |
| chr18_-_6634424 | 2.12 |
ENSDART00000062423
ENSDART00000179955 |
tnni1c
|
troponin I, skeletal, slow c |
| chr19_-_2317558 | 2.10 |
ENSDART00000190300
|
sp8a
|
sp8 transcription factor a |
| chr17_-_21066075 | 2.08 |
ENSDART00000078763
ENSDART00000104327 |
vsx1
|
visual system homeobox 1 homolog, chx10-like |
| chr7_+_7048245 | 2.06 |
ENSDART00000001649
|
actn3b
|
actinin alpha 3b |
| chr14_+_6423973 | 2.05 |
ENSDART00000051556
|
abca1b
|
ATP-binding cassette, sub-family A (ABC1), member 1B |
| chr6_+_23057311 | 2.03 |
ENSDART00000026448
|
evpla
|
envoplakin a |
| chr11_+_14622379 | 2.02 |
ENSDART00000112589
|
efna2b
|
ephrin-A2b |
| chr15_-_29387446 | 2.02 |
ENSDART00000145976
ENSDART00000035096 |
serpinh1b
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1b |
| chr2_-_44255537 | 2.01 |
ENSDART00000011188
ENSDART00000093298 |
atp1a2a
|
ATPase Na+/K+ transporting subunit alpha 2 |
| chr22_+_11775269 | 1.98 |
ENSDART00000140272
|
krt96
|
keratin 96 |
| chr3_-_28075756 | 1.92 |
ENSDART00000122037
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr15_-_15357178 | 1.92 |
ENSDART00000106120
|
ywhag2
|
3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide 2 |
| chr19_+_1184878 | 1.92 |
ENSDART00000163539
|
scrt1a
|
scratch family zinc finger 1a |
| chr23_+_35714574 | 1.88 |
ENSDART00000164616
|
tuba1c
|
tubulin, alpha 1c |
| chr7_+_31879649 | 1.86 |
ENSDART00000099789
|
mybpc3
|
myosin binding protein C, cardiac |
| chr15_+_32727848 | 1.86 |
ENSDART00000161361
|
postnb
|
periostin, osteoblast specific factor b |
| chr6_+_13742899 | 1.85 |
ENSDART00000104722
|
cdk5r2a
|
cyclin-dependent kinase 5, regulatory subunit 2a (p39) |
| chr22_+_11756040 | 1.83 |
ENSDART00000105808
|
krt97
|
keratin 97 |
| chr2_+_4208323 | 1.82 |
ENSDART00000167906
|
gata6
|
GATA binding protein 6 |
| chr11_-_18705303 | 1.82 |
ENSDART00000059732
|
id1
|
inhibitor of DNA binding 1 |
| chr14_+_7939398 | 1.77 |
ENSDART00000189773
|
cxxc5b
|
CXXC finger protein 5b |
| chr4_+_12615836 | 1.70 |
ENSDART00000003583
|
lmo3
|
LIM domain only 3 |
| chr2_-_42128714 | 1.68 |
ENSDART00000047055
|
trim55a
|
tripartite motif containing 55a |
| chr7_+_31879986 | 1.66 |
ENSDART00000138491
|
mybpc3
|
myosin binding protein C, cardiac |
| chr14_+_7939216 | 1.64 |
ENSDART00000171657
|
cxxc5b
|
CXXC finger protein 5b |
| chr10_+_15777064 | 1.64 |
ENSDART00000114483
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr4_+_7508316 | 1.64 |
ENSDART00000170924
ENSDART00000170933 ENSDART00000164985 ENSDART00000167571 ENSDART00000158843 ENSDART00000158999 |
tnnt2e
|
troponin T2e, cardiac |
| chr24_+_24461558 | 1.62 |
ENSDART00000182424
|
bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr7_+_6652967 | 1.62 |
ENSDART00000102681
|
pnp5a
|
purine nucleoside phosphorylase 5a |
| chr16_+_40575742 | 1.62 |
ENSDART00000161503
|
ccne2
|
cyclin E2 |
| chr3_-_36115339 | 1.61 |
ENSDART00000187406
ENSDART00000123505 ENSDART00000151775 |
rab11fip4a
|
RAB11 family interacting protein 4 (class II) a |
| chr1_+_26356205 | 1.59 |
ENSDART00000190064
ENSDART00000176380 |
tet2
|
tet methylcytosine dioxygenase 2 |
| chr23_+_37458602 | 1.55 |
ENSDART00000181686
|
cdaa
|
cytidine deaminase a |
| chr10_+_15777258 | 1.53 |
ENSDART00000140511
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr16_+_28728347 | 1.52 |
ENSDART00000149240
|
si:dkey-24i24.3
|
si:dkey-24i24.3 |
| chr20_+_35382482 | 1.52 |
ENSDART00000135284
|
vsnl1a
|
visinin-like 1a |
| chr5_+_37854685 | 1.51 |
ENSDART00000051222
ENSDART00000185283 |
ins
|
preproinsulin |
| chr1_+_25801648 | 1.51 |
ENSDART00000129471
|
gucy1b1
|
guanylate cyclase 1 soluble subunit beta 1 |
| chr9_+_23665777 | 1.49 |
ENSDART00000060905
|
gypc
|
glycophorin C (Gerbich blood group) |
| chr16_-_26676685 | 1.47 |
ENSDART00000103431
|
esrp1
|
epithelial splicing regulatory protein 1 |
| chr19_-_9711472 | 1.47 |
ENSDART00000016197
ENSDART00000175075 |
slc2a3a
|
solute carrier family 2 (facilitated glucose transporter), member 3a |
| chr1_-_25177086 | 1.47 |
ENSDART00000144711
ENSDART00000177225 |
tmem154
|
transmembrane protein 154 |
| chr15_+_28202170 | 1.45 |
ENSDART00000077736
|
vtna
|
vitronectin a |
| chr12_-_31103187 | 1.45 |
ENSDART00000005562
ENSDART00000031408 ENSDART00000125046 ENSDART00000009237 ENSDART00000122972 ENSDART00000153068 |
tcf7l2
|
transcription factor 7 like 2 |
| chr3_+_23654233 | 1.39 |
ENSDART00000078428
|
hoxb13a
|
homeobox B13a |
| chr8_-_14151051 | 1.37 |
ENSDART00000090427
|
si:dkey-6n6.1
|
si:dkey-6n6.1 |
| chr13_+_24834199 | 1.37 |
ENSDART00000101274
|
zgc:153981
|
zgc:153981 |
| chr21_+_5974590 | 1.37 |
ENSDART00000098499
|
dck
|
deoxycytidine kinase |
| chr6_+_23887314 | 1.36 |
ENSDART00000163188
|
znf648
|
zinc finger protein 648 |
| chr8_-_50981175 | 1.33 |
ENSDART00000004065
|
zgc:91909
|
zgc:91909 |
| chr13_+_1575276 | 1.32 |
ENSDART00000165987
|
DST
|
dystonin |
| chr15_+_1397811 | 1.29 |
ENSDART00000102125
|
schip1
|
schwannomin interacting protein 1 |
| chr5_-_17601759 | 1.28 |
ENSDART00000138387
|
si:ch211-130h14.6
|
si:ch211-130h14.6 |
| chr23_-_24263474 | 1.26 |
ENSDART00000160312
|
hspb7
|
heat shock protein family, member 7 (cardiovascular) |
| chr24_+_20575259 | 1.26 |
ENSDART00000010488
|
klhl40b
|
kelch-like family member 40b |
| chr19_+_9174166 | 1.25 |
ENSDART00000104637
ENSDART00000150968 |
si:ch211-81a5.8
|
si:ch211-81a5.8 |
| chr6_-_47838481 | 1.25 |
ENSDART00000185284
|
lrig2
|
leucine-rich repeats and immunoglobulin-like domains 2 |
| chr6_-_8865217 | 1.24 |
ENSDART00000151443
|
tmeff2b
|
transmembrane protein with EGF-like and two follistatin-like domains 2b |
| chr14_-_32255126 | 1.23 |
ENSDART00000017259
|
fgf13a
|
fibroblast growth factor 13a |
| chr7_+_10610791 | 1.21 |
ENSDART00000166064
|
fah
|
fumarylacetoacetate hydrolase (fumarylacetoacetase) |
| chr23_-_31346319 | 1.21 |
ENSDART00000008401
ENSDART00000138106 |
phip
|
pleckstrin homology domain interacting protein |
| chr15_-_39969988 | 1.19 |
ENSDART00000146054
|
rps5
|
ribosomal protein S5 |
| chr17_+_6276559 | 1.19 |
ENSDART00000131075
|
dusp23b
|
dual specificity phosphatase 23b |
| chr7_+_26224211 | 1.17 |
ENSDART00000173999
|
vgf
|
VGF nerve growth factor inducible |
| chr1_-_40911332 | 1.15 |
ENSDART00000027463
|
hmx4
|
H6 family homeobox 4 |
| chr13_-_31452516 | 1.15 |
ENSDART00000193268
|
rtn1a
|
reticulon 1a |
| chr1_-_17587552 | 1.14 |
ENSDART00000039917
|
acsl1a
|
acyl-CoA synthetase long chain family member 1a |
| chr10_+_26667475 | 1.14 |
ENSDART00000133281
ENSDART00000147013 |
si:ch73-52f15.5
|
si:ch73-52f15.5 |
| chr3_+_20156956 | 1.12 |
ENSDART00000125281
|
ngfra
|
nerve growth factor receptor a (TNFR superfamily, member 16) |
| chr13_-_12581388 | 1.12 |
ENSDART00000079655
|
enpep
|
glutamyl aminopeptidase |
| chr23_+_23658474 | 1.12 |
ENSDART00000162838
|
agrn
|
agrin |
| chr1_-_20593778 | 1.12 |
ENSDART00000124770
|
ugt8
|
UDP glycosyltransferase 8 |
| chr21_+_11468934 | 1.09 |
ENSDART00000126045
ENSDART00000129744 ENSDART00000102368 |
grin1a
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1a |
| chr3_+_17910569 | 1.08 |
ENSDART00000080946
|
ttc25
|
tetratricopeptide repeat domain 25 |
| chr12_-_28570989 | 1.07 |
ENSDART00000008010
|
pdk2a
|
pyruvate dehydrogenase kinase, isozyme 2a |
| chr14_-_41678357 | 1.07 |
ENSDART00000185925
|
fgfrl1b
|
fibroblast growth factor receptor like 1b |
| chr22_+_465269 | 1.05 |
ENSDART00000145767
|
celsr2
|
cadherin, EGF LAG seven-pass G-type receptor 2 |
| chr6_-_40098641 | 1.05 |
ENSDART00000017402
|
ip6k2b
|
inositol hexakisphosphate kinase 2b |
| chr17_-_51195651 | 1.04 |
ENSDART00000191205
ENSDART00000088185 |
paplna
|
papilin a, proteoglycan-like sulfated glycoprotein |
| chr8_+_22289320 | 1.02 |
ENSDART00000075126
|
b3gnt7l
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 7, like |
| chr18_-_14337450 | 1.01 |
ENSDART00000061435
|
hsbp1b
|
heat shock factor binding protein 1b |
| chr7_+_27277236 | 1.00 |
ENSDART00000185161
|
sox6
|
SRY (sex determining region Y)-box 6 |
| chr23_-_27647769 | 0.98 |
ENSDART00000059974
|
wnt10b
|
wingless-type MMTV integration site family, member 10b |
| chr5_+_42097608 | 0.97 |
ENSDART00000014404
|
p2rx5
|
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr18_+_3052408 | 0.96 |
ENSDART00000181563
|
kctd14
|
potassium channel tetramerization domain containing 14 |
| chr5_-_51819027 | 0.96 |
ENSDART00000164267
|
homer1b
|
homer scaffolding protein 1b |
| chr5_+_3501859 | 0.96 |
ENSDART00000080486
|
ywhag1
|
3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide 1 |
| chr13_+_18545819 | 0.95 |
ENSDART00000101859
ENSDART00000110197 ENSDART00000136064 |
zgc:154058
|
zgc:154058 |
| chr21_+_11468642 | 0.94 |
ENSDART00000041869
|
grin1a
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1a |
| chr6_-_8865543 | 0.92 |
ENSDART00000042115
|
tmeff2b
|
transmembrane protein with EGF-like and two follistatin-like domains 2b |
| chr2_-_30659222 | 0.91 |
ENSDART00000145405
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr17_+_38262408 | 0.91 |
ENSDART00000017493
|
nkx2.1
|
NK2 homeobox 1 |
| chr8_+_7033049 | 0.91 |
ENSDART00000064172
ENSDART00000134440 |
gpd1a
|
glycerol-3-phosphate dehydrogenase 1a |
| chr25_+_36674715 | 0.90 |
ENSDART00000111861
|
mafb
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog b (paralog b) |
| chr12_-_28818720 | 0.90 |
ENSDART00000134453
ENSDART00000141727 |
prr15lb
|
proline rich 15-like b |
| chr21_+_13205859 | 0.90 |
ENSDART00000102253
|
sptan1
|
spectrin alpha, non-erythrocytic 1 |
| chr20_-_42439896 | 0.89 |
ENSDART00000061049
|
vgll2a
|
vestigial-like family member 2a |
| chr4_+_12617108 | 0.88 |
ENSDART00000134362
ENSDART00000112860 |
lmo3
|
LIM domain only 3 |
| chr5_+_34622320 | 0.86 |
ENSDART00000141338
|
enc1
|
ectodermal-neural cortex 1 |
| chr8_-_11229523 | 0.86 |
ENSDART00000002164
|
unc45b
|
unc-45 myosin chaperone B |
| chr11_-_25384213 | 0.86 |
ENSDART00000103650
|
mafbb
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Bb |
| chr7_+_63325819 | 0.85 |
ENSDART00000085612
ENSDART00000161436 |
pcdh7b
|
protocadherin 7b |
| chr4_-_75175407 | 0.85 |
ENSDART00000180125
|
CABZ01043953.1
|
|
| chr25_-_36044583 | 0.84 |
ENSDART00000073432
|
rbl2
|
retinoblastoma-like 2 (p130) |
| chr21_-_20929575 | 0.84 |
ENSDART00000163889
|
c6
|
complement component 6 |
| chr18_-_20560007 | 0.84 |
ENSDART00000141367
ENSDART00000090186 |
si:ch211-238n5.4
|
si:ch211-238n5.4 |
| chr10_+_25355308 | 0.84 |
ENSDART00000100415
|
map3k7cl
|
map3k7 C-terminal like |
| chr21_-_17482465 | 0.84 |
ENSDART00000004548
|
barhl1b
|
BarH-like homeobox 1b |
| chr15_+_9327252 | 0.84 |
ENSDART00000144381
|
sgcg
|
sarcoglycan, gamma |
| chr5_-_60159116 | 0.83 |
ENSDART00000147675
|
si:dkey-280e8.1
|
si:dkey-280e8.1 |
| chr6_+_23931236 | 0.83 |
ENSDART00000166079
|
gadd45ab
|
growth arrest and DNA-damage-inducible, alpha, b |
| chr18_+_48802154 | 0.80 |
ENSDART00000191403
|
bmp16
|
bone morphogenetic protein 16 |
| chr19_-_32150078 | 0.79 |
ENSDART00000134934
ENSDART00000186410 ENSDART00000181780 |
pag1
|
phosphoprotein membrane anchor with glycosphingolipid microdomains 1 |
| chr8_+_14381272 | 0.79 |
ENSDART00000057642
|
acbd6
|
acyl-CoA binding domain containing 6 |
| chr13_+_43400443 | 0.79 |
ENSDART00000084321
|
dact2
|
dishevelled-binding antagonist of beta-catenin 2 |
| chr9_-_98982 | 0.78 |
ENSDART00000147882
|
lims2
|
LIM and senescent cell antigen-like domains 2 |
| chr7_+_49664174 | 0.77 |
ENSDART00000137059
ENSDART00000131210 |
rassf7b
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7b |
| chr25_-_35542739 | 0.76 |
ENSDART00000097651
|
si:ch211-87j1.4
|
si:ch211-87j1.4 |
| chr14_+_30730749 | 0.76 |
ENSDART00000087884
|
ccdc85b
|
coiled-coil domain containing 85B |
| chr10_-_24371312 | 0.75 |
ENSDART00000149362
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr1_-_26293203 | 0.75 |
ENSDART00000180140
|
cxxc4
|
CXXC finger 4 |
| chr18_+_35742838 | 0.74 |
ENSDART00000088504
ENSDART00000140386 |
rasgrp4
|
RAS guanyl releasing protein 4 |
| chr5_+_4806851 | 0.73 |
ENSDART00000067599
|
angptl2a
|
angiopoietin-like 2a |
| chr13_+_24287093 | 0.73 |
ENSDART00000058628
|
ccsapb
|
centriole, cilia and spindle-associated protein b |
| chr5_-_10768258 | 0.73 |
ENSDART00000157043
|
rtn4r
|
reticulon 4 receptor |
| chr13_-_7575216 | 0.72 |
ENSDART00000159443
|
pitx3
|
paired-like homeodomain 3 |
| chr7_-_20836625 | 0.72 |
ENSDART00000192566
|
cldn15a
|
claudin 15a |
| chr22_-_18179214 | 0.71 |
ENSDART00000129576
|
si:ch211-125m10.6
|
si:ch211-125m10.6 |
| chr20_-_26467307 | 0.70 |
ENSDART00000078072
ENSDART00000158213 |
akap12b
|
A kinase (PRKA) anchor protein 12b |
| chr6_+_4872883 | 0.70 |
ENSDART00000186730
ENSDART00000092290 ENSDART00000151674 |
pcdh9
|
protocadherin 9 |
| chr7_-_35516251 | 0.69 |
ENSDART00000045628
|
irx6a
|
iroquois homeobox 6a |
| chr1_-_17650223 | 0.69 |
ENSDART00000043484
|
si:dkey-256e7.5
|
si:dkey-256e7.5 |
| chr4_+_8532580 | 0.69 |
ENSDART00000162065
|
wnt5b
|
wingless-type MMTV integration site family, member 5b |
| chr7_-_68373495 | 0.67 |
ENSDART00000167440
|
zfhx3
|
zinc finger homeobox 3 |
| chr7_-_23745984 | 0.66 |
ENSDART00000048050
|
ITGB1BP2
|
zgc:92429 |
| chr12_+_30360184 | 0.66 |
ENSDART00000190718
|
si:ch211-225b10.3
|
si:ch211-225b10.3 |
| chr8_+_21376290 | 0.64 |
ENSDART00000136765
|
ela2
|
elastase 2 |
| chr19_-_31341850 | 0.64 |
ENSDART00000040810
|
arl4ab
|
ADP-ribosylation factor-like 4ab |
| chr12_+_17504559 | 0.64 |
ENSDART00000020628
|
cyth3a
|
cytohesin 3a |
| chr12_+_30360579 | 0.64 |
ENSDART00000152900
|
si:ch211-225b10.3
|
si:ch211-225b10.3 |
| chr8_+_50727220 | 0.63 |
ENSDART00000127062
|
egr3
|
early growth response 3 |
| chr6_-_31224563 | 0.63 |
ENSDART00000104616
|
lepr
|
leptin receptor |
| chr18_-_10995410 | 0.62 |
ENSDART00000136751
|
tspan33b
|
tetraspanin 33b |
| chr4_-_23858900 | 0.62 |
ENSDART00000123199
|
usp6nl
|
USP6 N-terminal like |
| chr2_+_32016256 | 0.61 |
ENSDART00000005143
|
mycb
|
MYC proto-oncogene, bHLH transcription factor b |
| chr20_+_27020201 | 0.61 |
ENSDART00000126919
ENSDART00000016014 |
chga
|
chromogranin A |
| chr7_+_27253063 | 0.61 |
ENSDART00000191138
|
sox6
|
SRY (sex determining region Y)-box 6 |
| chr9_-_53666031 | 0.59 |
ENSDART00000126314
|
pcdh8
|
protocadherin 8 |
| chr16_+_40560622 | 0.58 |
ENSDART00000038294
|
tp53inp1
|
tumor protein p53 inducible nuclear protein 1 |
| chr6_+_28051978 | 0.58 |
ENSDART00000143218
|
si:ch73-194h10.2
|
si:ch73-194h10.2 |
| chr18_-_14337065 | 0.58 |
ENSDART00000135703
|
hsbp1b
|
heat shock factor binding protein 1b |
| chr23_-_8373676 | 0.56 |
ENSDART00000105135
ENSDART00000158531 |
oprl1
|
opiate receptor-like 1 |
| chr7_+_24251367 | 0.56 |
ENSDART00000173642
|
svep1
|
sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 |
| chr22_+_7486867 | 0.56 |
ENSDART00000034586
|
CELA1 (1 of many)
|
zgc:112302 |
| chr7_+_20471315 | 0.56 |
ENSDART00000173714
|
si:dkey-19b23.13
|
si:dkey-19b23.13 |
| chr1_-_26292897 | 0.56 |
ENSDART00000112899
ENSDART00000185410 |
cxxc4
|
CXXC finger 4 |
| chr10_+_34315719 | 0.56 |
ENSDART00000135303
|
stard13a
|
StAR-related lipid transfer (START) domain containing 13a |
| chr9_-_8454060 | 0.55 |
ENSDART00000110158
|
irs2b
|
insulin receptor substrate 2b |
| chr24_+_38155830 | 0.54 |
ENSDART00000152019
|
si:ch211-234p6.5
|
si:ch211-234p6.5 |
| chr15_-_18176694 | 0.54 |
ENSDART00000189840
|
tmprss5
|
transmembrane protease, serine 5 |
| chr5_-_31904562 | 0.54 |
ENSDART00000140640
|
coro1cb
|
coronin, actin binding protein, 1Cb |
| chr20_+_20902549 | 0.53 |
ENSDART00000181195
|
brf1b
|
BRF1, RNA polymerase III transcription initiation factor b |
| chr4_+_20255160 | 0.53 |
ENSDART00000188658
|
lrtm2a
|
leucine-rich repeats and transmembrane domains 2a |
| chr1_+_27825980 | 0.53 |
ENSDART00000160524
|
pspc1
|
paraspeckle component 1 |
| chr1_-_25911292 | 0.52 |
ENSDART00000145012
|
usp53b
|
ubiquitin specific peptidase 53b |
| chr16_+_3004422 | 0.52 |
ENSDART00000189969
|
CABZ01043952.1
|
|
| chr18_+_19820008 | 0.51 |
ENSDART00000079691
ENSDART00000135049 |
iqch
|
IQ motif containing H |
| chr6_+_39222598 | 0.51 |
ENSDART00000154991
|
b4galnt1b
|
beta-1,4-N-acetyl-galactosaminyl transferase 1b |
| chr16_-_14353567 | 0.51 |
ENSDART00000139859
|
itga10
|
integrin, alpha 10 |
| chr22_+_35472653 | 0.51 |
ENSDART00000076424
ENSDART00000187204 |
tctex1d2
|
Tctex1 domain containing 2 |
| chr1_+_2101541 | 0.49 |
ENSDART00000128187
ENSDART00000167050 ENSDART00000182153 ENSDART00000122626 ENSDART00000164488 |
mbnl2
|
muscleblind-like splicing regulator 2 |
| chr14_+_26805388 | 0.49 |
ENSDART00000044389
ENSDART00000173126 |
klhl4
|
kelch-like family member 4 |
| chr7_-_38638276 | 0.49 |
ENSDART00000074463
|
c6ast4
|
six-cysteine containing astacin protease 4 |
| chr16_+_46401576 | 0.49 |
ENSDART00000130264
|
rpz
|
rapunzel |
| chr8_-_30242706 | 0.47 |
ENSDART00000139864
ENSDART00000143809 |
zgc:162939
|
zgc:162939 |
| chr13_+_40770628 | 0.46 |
ENSDART00000085846
|
nkx1.2la
|
NK1 transcription factor related 2-like,a |
| chr4_+_20263097 | 0.46 |
ENSDART00000138820
|
lrtm2a
|
leucine-rich repeats and transmembrane domains 2a |
| chr1_+_33401774 | 0.45 |
ENSDART00000149786
ENSDART00000110026 ENSDART00000148946 |
dhrsx
|
dehydrogenase/reductase (SDR family) X-linked |
| chr17_+_53156530 | 0.45 |
ENSDART00000126277
ENSDART00000156774 |
dph6
|
diphthamine biosynthesis 6 |
| chr24_-_35707552 | 0.45 |
ENSDART00000165199
|
mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr21_-_14310159 | 0.45 |
ENSDART00000155097
|
si:ch211-196i2.1
|
si:ch211-196i2.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of msc
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:0006600 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 0.7 | 3.5 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.7 | 2.1 | GO:0048340 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) |
| 0.7 | 2.0 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.6 | 2.4 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.5 | 3.1 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.5 | 1.6 | GO:0009972 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.4 | 1.3 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.4 | 1.5 | GO:0048521 | positive regulation of epithelial cell differentiation(GO:0030858) negative regulation of behavior(GO:0048521) negative regulation of feeding behavior(GO:2000252) |
| 0.4 | 1.5 | GO:0099543 | trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.4 | 1.1 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.4 | 1.8 | GO:0043011 | myeloid dendritic cell activation(GO:0001773) myeloid dendritic cell differentiation(GO:0043011) |
| 0.4 | 1.4 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.4 | 2.1 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.3 | 2.4 | GO:0098773 | skin epidermis development(GO:0098773) |
| 0.3 | 2.6 | GO:0006662 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.3 | 2.2 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.3 | 2.1 | GO:0033700 | phospholipid efflux(GO:0033700) |
| 0.2 | 2.4 | GO:0031268 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.2 | 0.7 | GO:0032060 | bleb assembly(GO:0032060) slow muscle cell migration(GO:1904969) |
| 0.2 | 1.6 | GO:0006211 | 5-methylcytosine catabolic process(GO:0006211) 5-methylcytosine metabolic process(GO:0019857) |
| 0.2 | 0.9 | GO:0046168 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.2 | 1.3 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.2 | 1.3 | GO:0003319 | cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) |
| 0.2 | 0.6 | GO:0090278 | negative regulation of peptide secretion(GO:0002792) negative regulation of peptide hormone secretion(GO:0090278) |
| 0.2 | 1.2 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.2 | 0.7 | GO:0060295 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.2 | 2.9 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.2 | 1.6 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.1 | 0.7 | GO:0008591 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.1 | 1.0 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 0.8 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.1 | 2.4 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.1 | 2.3 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.1 | 2.2 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.1 | 1.9 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.1 | 2.2 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 2.0 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.1 | 1.1 | GO:0030656 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.1 | 1.7 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 | 0.8 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 1.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 1.1 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.1 | 2.2 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.1 | 0.2 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 1.0 | GO:0033198 | response to ATP(GO:0033198) |
| 0.1 | 0.4 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 0.6 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.1 | 0.9 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 2.8 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.1 | 0.4 | GO:0042403 | thyroid hormone metabolic process(GO:0042403) |
| 0.1 | 0.4 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.1 | 0.2 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.1 | 2.4 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.1 | 2.4 | GO:0035567 | non-canonical Wnt signaling pathway(GO:0035567) |
| 0.1 | 0.4 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 0.4 | GO:1902221 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.1 | 1.2 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 2.0 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 0.6 | GO:0019233 | sensory perception of pain(GO:0019233) |
| 0.1 | 0.9 | GO:0001843 | neural tube closure(GO:0001843) |
| 0.0 | 0.4 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.4 | GO:0006337 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 1.1 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.4 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.0 | 2.0 | GO:0045103 | intermediate filament-based process(GO:0045103) intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 1.8 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 1.1 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.3 | GO:0048260 | positive regulation of receptor-mediated endocytosis(GO:0048260) protein localization to early endosome(GO:1902946) |
| 0.0 | 1.6 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 1.6 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.0 | 2.1 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 1.8 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.8 | GO:2000134 | negative regulation of cell cycle G1/S phase transition(GO:1902807) negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.5 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.1 | GO:0046689 | response to mercury ion(GO:0046689) detoxification of mercury ion(GO:0050787) |
| 0.0 | 0.5 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.4 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.2 | GO:0070291 | N-acylethanolamine metabolic process(GO:0070291) |
| 0.0 | 0.7 | GO:0021984 | adenohypophysis development(GO:0021984) |
| 0.0 | 0.7 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 1.6 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 1.2 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 1.8 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.8 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.8 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.2 | GO:0072091 | forebrain anterior/posterior pattern specification(GO:0021797) regulation of stem cell proliferation(GO:0072091) |
| 0.0 | 1.5 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.0 | 0.6 | GO:0009749 | response to glucose(GO:0009749) |
| 0.0 | 1.1 | GO:0010675 | regulation of cellular carbohydrate metabolic process(GO:0010675) regulation of glucose metabolic process(GO:0010906) |
| 0.0 | 0.7 | GO:0035148 | tube formation(GO:0035148) |
| 0.0 | 0.2 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.5 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 1.3 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 1.0 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.5 | GO:0042752 | regulation of circadian rhythm(GO:0042752) |
| 0.0 | 0.8 | GO:0050868 | negative regulation of T cell activation(GO:0050868) negative regulation of leukocyte cell-cell adhesion(GO:1903038) |
| 0.0 | 0.4 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.3 | GO:0050926 | regulation of positive chemotaxis(GO:0050926) positive regulation of positive chemotaxis(GO:0050927) induction of positive chemotaxis(GO:0050930) |
| 0.0 | 1.0 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.5 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.3 | GO:0030534 | adult behavior(GO:0030534) |
| 0.0 | 0.9 | GO:0001764 | neuron migration(GO:0001764) |
| 0.0 | 1.0 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.4 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.8 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.2 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 1.5 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 1.9 | GO:0060047 | heart contraction(GO:0060047) |
| 0.0 | 0.9 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.2 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.4 | 1.8 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.4 | 1.4 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.3 | 0.9 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.2 | 2.4 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.2 | 2.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.2 | 0.7 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.2 | 0.9 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 0.8 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 2.2 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.1 | 1.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 3.8 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.5 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.1 | 0.6 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 5.8 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 0.8 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 1.3 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 1.4 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 1.6 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 2.4 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 2.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.7 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 1.2 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 1.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 2.5 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 6.3 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 1.5 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.0 | 1.9 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.9 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.7 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.4 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.6 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.5 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.8 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 4.0 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.0 | GO:0005787 | signal peptidase complex(GO:0005787) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 4.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.3 | 1.4 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.3 | 2.0 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.3 | 4.7 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.3 | 3.5 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.2 | 1.2 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.2 | 1.6 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.2 | 0.9 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.2 | 1.6 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.2 | 2.0 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.2 | 1.4 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.2 | 1.0 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.2 | 1.6 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.2 | 1.1 | GO:0005035 | death receptor activity(GO:0005035) |
| 0.1 | 1.6 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.1 | 2.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 1.8 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 1.8 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 1.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 3.2 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 2.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 2.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.8 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 1.7 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 1.1 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 2.4 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 2.0 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.4 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) NADH binding(GO:0070404) |
| 0.1 | 1.5 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 1.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 1.0 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 2.4 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 0.5 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.1 | 1.1 | GO:0043236 | laminin binding(GO:0043236) |
| 0.1 | 0.9 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 1.4 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 1.6 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.1 | 0.5 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 0.3 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 0.8 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 0.6 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 0.4 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 1.4 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 1.8 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.3 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 1.2 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 1.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.2 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 1.1 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 1.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 1.6 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 2.2 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.5 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 1.2 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.8 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 1.2 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 1.2 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.7 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.6 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.9 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 2.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.6 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 1.4 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.7 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 1.3 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 16.0 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.4 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 1.5 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.3 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 2.6 | GO:0008757 | S-adenosylmethionine-dependent methyltransferase activity(GO:0008757) |
| 0.0 | 0.2 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 2.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 2.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 1.6 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 0.8 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 1.8 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 1.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 2.4 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 1.3 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.7 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.4 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.8 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 3.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.8 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.5 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.3 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.6 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.7 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.9 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.4 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.4 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.6 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.3 | 2.4 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.3 | 1.5 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.2 | 2.7 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 1.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 1.9 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 0.8 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 0.9 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 0.3 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.1 | 1.1 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 0.4 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.1 | 3.3 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 1.1 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.8 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 1.2 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 3.5 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.8 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.3 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |