Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
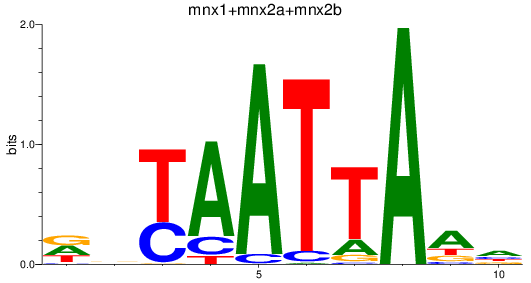
Results for mnx1+mnx2a+mnx2b
Z-value: 0.40
Transcription factors associated with mnx1+mnx2a+mnx2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mnx2b
|
ENSDARG00000030350 | motor neuron and pancreas homeobox 2b |
|
mnx1
|
ENSDARG00000035984 | motor neuron and pancreas homeobox 1 |
|
mnx2a
|
ENSDARG00000042106 | motor neuron and pancreas homeobox 2a |
|
mnx1
|
ENSDARG00000109419 | motor neuron and pancreas homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mnx1 | dr11_v1_chr7_+_40638210_40638210 | -0.74 | 2.3e-02 | Click! |
| mnx2a | dr11_v1_chr9_+_7724152_7724152 | -0.72 | 3.0e-02 | Click! |
| mnx2b | dr11_v1_chr1_-_5455498_5455502 | -0.55 | 1.2e-01 | Click! |
Activity profile of mnx1+mnx2a+mnx2b motif
Sorted Z-values of mnx1+mnx2a+mnx2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_45334255 | 0.90 |
ENSDART00000126848
ENSDART00000134161 ENSDART00000142322 ENSDART00000145011 ENSDART00000183560 |
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr16_-_29387215 | 0.71 |
ENSDART00000148787
|
s100a1
|
S100 calcium binding protein A1 |
| chr10_-_21362071 | 0.69 |
ENSDART00000125167
|
avd
|
avidin |
| chr10_-_21362320 | 0.67 |
ENSDART00000189789
|
avd
|
avidin |
| chr10_-_25217347 | 0.62 |
ENSDART00000036906
|
kpna7
|
karyopherin alpha 7 (importin alpha 8) |
| chr11_-_44801968 | 0.61 |
ENSDART00000161846
|
map1lc3c
|
microtubule-associated protein 1 light chain 3 gamma |
| chr11_-_6452444 | 0.55 |
ENSDART00000137879
ENSDART00000134957 ENSDART00000004483 |
larp6b
|
La ribonucleoprotein domain family, member 6b |
| chr11_+_18157260 | 0.54 |
ENSDART00000144659
|
zgc:173545
|
zgc:173545 |
| chr11_+_18130300 | 0.53 |
ENSDART00000169146
|
zgc:175135
|
zgc:175135 |
| chr8_+_41037541 | 0.49 |
ENSDART00000129344
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr20_-_34028967 | 0.48 |
ENSDART00000153408
ENSDART00000033817 |
scyl3
|
SCY1-like, kinase-like 3 |
| chr14_+_34490445 | 0.47 |
ENSDART00000132193
ENSDART00000148044 |
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr19_+_42432625 | 0.44 |
ENSDART00000076938
|
pogza
|
pogo transposable element derived with ZNF domain a |
| chr10_+_6884627 | 0.44 |
ENSDART00000125262
ENSDART00000121729 ENSDART00000105384 |
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr16_+_39159752 | 0.42 |
ENSDART00000122081
|
sybu
|
syntabulin (syntaxin-interacting) |
| chr20_-_23426339 | 0.41 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr17_+_43867889 | 0.40 |
ENSDART00000132673
ENSDART00000167214 |
zgc:66313
|
zgc:66313 |
| chr5_-_48664522 | 0.38 |
ENSDART00000083229
|
mblac2
|
metallo-beta-lactamase domain containing 2 |
| chr17_-_40956035 | 0.38 |
ENSDART00000124715
|
si:dkey-16j16.4
|
si:dkey-16j16.4 |
| chr23_+_45200481 | 0.37 |
ENSDART00000004357
ENSDART00000111126 ENSDART00000193560 ENSDART00000190476 |
psip1b
|
PC4 and SFRS1 interacting protein 1b |
| chr14_+_35405518 | 0.36 |
ENSDART00000171565
|
zbtb3
|
zinc finger and BTB domain containing 3 |
| chr17_+_16090436 | 0.36 |
ENSDART00000136059
ENSDART00000138734 |
znf395a
|
zinc finger protein 395a |
| chr14_+_35428152 | 0.36 |
ENSDART00000172597
|
sytl4
|
synaptotagmin-like 4 |
| chr17_+_14965570 | 0.34 |
ENSDART00000066604
|
gpr137c
|
G protein-coupled receptor 137c |
| chr16_-_42965192 | 0.34 |
ENSDART00000113714
|
mtx1a
|
metaxin 1a |
| chr19_+_2631565 | 0.34 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr13_-_25719628 | 0.34 |
ENSDART00000135383
|
si:dkey-192p21.6
|
si:dkey-192p21.6 |
| chr10_-_8046764 | 0.33 |
ENSDART00000099031
|
zgc:136254
|
zgc:136254 |
| chr16_-_42056137 | 0.32 |
ENSDART00000102798
|
zp3d.2
|
zona pellucida glycoprotein 3d tandem duplicate 2 |
| chr3_-_32873641 | 0.31 |
ENSDART00000075277
|
zgc:113090
|
zgc:113090 |
| chr20_-_37813863 | 0.31 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr10_+_33744098 | 0.30 |
ENSDART00000147775
|
rxfp2a
|
relaxin/insulin-like family peptide receptor 2a |
| chr1_+_35985813 | 0.30 |
ENSDART00000179634
ENSDART00000139636 ENSDART00000175902 |
zgc:152968
|
zgc:152968 |
| chr22_-_24818066 | 0.30 |
ENSDART00000143443
|
vtg6
|
vitellogenin 6 |
| chr9_-_38021889 | 0.29 |
ENSDART00000183482
ENSDART00000124333 |
adcy5
|
adenylate cyclase 5 |
| chr13_-_27354003 | 0.28 |
ENSDART00000101479
ENSDART00000044652 |
ddx43
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 43 |
| chr1_+_44173245 | 0.28 |
ENSDART00000159450
ENSDART00000106048 ENSDART00000157763 |
ctnnd1
|
catenin (cadherin-associated protein), delta 1 |
| chr10_-_8053385 | 0.27 |
ENSDART00000142714
|
si:ch211-251f6.7
|
si:ch211-251f6.7 |
| chr14_+_16151368 | 0.27 |
ENSDART00000160973
|
polr1a
|
polymerase (RNA) I polypeptide A |
| chr6_-_43283122 | 0.26 |
ENSDART00000186022
|
frmd4ba
|
FERM domain containing 4Ba |
| chr11_-_39118882 | 0.26 |
ENSDART00000113185
ENSDART00000156526 |
ap5b1
|
adaptor-related protein complex 5, beta 1 subunit |
| chr20_-_45060241 | 0.26 |
ENSDART00000185227
|
klhl29
|
kelch-like family member 29 |
| chr13_-_4018888 | 0.26 |
ENSDART00000058238
|
tjap1
|
tight junction associated protein 1 (peripheral) |
| chr20_+_27712714 | 0.26 |
ENSDART00000008306
|
zbtb1
|
zinc finger and BTB domain containing 1 |
| chr21_-_3853204 | 0.26 |
ENSDART00000188829
|
st6galnac4
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 |
| chr14_+_16151636 | 0.25 |
ENSDART00000159352
|
polr1a
|
polymerase (RNA) I polypeptide A |
| chr15_+_31344472 | 0.25 |
ENSDART00000146695
ENSDART00000159182 ENSDART00000060125 |
or107-1
|
odorant receptor, family D, subfamily 107, member 1 |
| chr12_-_13549538 | 0.25 |
ENSDART00000133895
|
ghdc
|
GH3 domain containing |
| chr2_+_41526904 | 0.25 |
ENSDART00000127520
|
acvr1l
|
activin A receptor, type 1 like |
| chr11_+_31864921 | 0.25 |
ENSDART00000180252
|
diaph3
|
diaphanous-related formin 3 |
| chr19_+_15441022 | 0.24 |
ENSDART00000098970
ENSDART00000140276 |
lin28a
|
lin-28 homolog A (C. elegans) |
| chr5_-_13206878 | 0.24 |
ENSDART00000051666
|
ppm1f
|
protein phosphatase, Mg2+/Mn2+ dependent, 1F |
| chr8_-_19051906 | 0.23 |
ENSDART00000089024
|
sema6bb
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6Bb |
| chr22_-_10440688 | 0.23 |
ENSDART00000111962
|
nol8
|
nucleolar protein 8 |
| chr24_-_30862168 | 0.22 |
ENSDART00000168540
|
ptbp2a
|
polypyrimidine tract binding protein 2a |
| chr11_-_25257045 | 0.22 |
ENSDART00000130477
|
snai1a
|
snail family zinc finger 1a |
| chr17_+_49500820 | 0.22 |
ENSDART00000170306
|
AREL1
|
apoptosis resistant E3 ubiquitin protein ligase 1 |
| chr13_+_13945218 | 0.22 |
ENSDART00000089501
ENSDART00000142997 |
eif2ak3
|
eukaryotic translation initiation factor 2-alpha kinase 3 |
| chr8_-_39822917 | 0.22 |
ENSDART00000067843
|
zgc:162025
|
zgc:162025 |
| chr22_-_10397600 | 0.21 |
ENSDART00000181964
ENSDART00000142886 |
nisch
|
nischarin |
| chr10_-_28028998 | 0.21 |
ENSDART00000023545
ENSDART00000143487 |
ints2
|
integrator complex subunit 2 |
| chr13_+_38817871 | 0.20 |
ENSDART00000187708
|
col19a1
|
collagen, type XIX, alpha 1 |
| chr9_-_14273652 | 0.19 |
ENSDART00000135458
|
abcb6b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6b |
| chr15_+_25528290 | 0.19 |
ENSDART00000123143
|
npat
|
nuclear protein, ataxia-telangiectasia locus |
| chr8_+_25145464 | 0.19 |
ENSDART00000136505
|
gnai3
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3 |
| chr17_+_28103675 | 0.19 |
ENSDART00000188078
|
zgc:91908
|
zgc:91908 |
| chr21_+_45502621 | 0.19 |
ENSDART00000166719
|
si:dkey-223p19.2
|
si:dkey-223p19.2 |
| chr17_+_16046314 | 0.19 |
ENSDART00000154554
ENSDART00000154338 ENSDART00000155336 |
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr1_-_45616470 | 0.19 |
ENSDART00000150165
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr19_+_40069524 | 0.18 |
ENSDART00000151365
ENSDART00000140926 |
zmym4
|
zinc finger, MYM-type 4 |
| chr12_-_6880694 | 0.18 |
ENSDART00000171846
|
pcdh15b
|
protocadherin-related 15b |
| chr10_-_13343831 | 0.18 |
ENSDART00000135941
|
il11ra
|
interleukin 11 receptor, alpha |
| chr9_-_50001606 | 0.18 |
ENSDART00000161648
ENSDART00000168514 |
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr7_-_23768234 | 0.18 |
ENSDART00000173981
|
si:ch211-200p22.4
|
si:ch211-200p22.4 |
| chr7_-_71389375 | 0.18 |
ENSDART00000128928
|
CABZ01074298.1
|
|
| chr15_-_26931541 | 0.18 |
ENSDART00000027563
|
ccdc9
|
coiled-coil domain containing 9 |
| chr13_-_31017960 | 0.17 |
ENSDART00000145287
|
wdfy4
|
WDFY family member 4 |
| chr7_+_39418869 | 0.17 |
ENSDART00000169195
|
CT030188.1
|
|
| chr1_-_32111882 | 0.17 |
ENSDART00000112639
ENSDART00000101958 |
pnpla4
|
patatin-like phospholipase domain containing 4 |
| chr15_-_16177603 | 0.17 |
ENSDART00000156352
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| chr25_-_15268443 | 0.17 |
ENSDART00000151827
|
ccdc73
|
coiled-coil domain containing 73 |
| chr8_+_21229718 | 0.16 |
ENSDART00000100222
|
cry1ba
|
cryptochrome circadian clock 1ba |
| chr10_-_32494499 | 0.16 |
ENSDART00000129395
|
uvrag
|
UV radiation resistance associated gene |
| chr12_+_30367371 | 0.15 |
ENSDART00000153364
|
ccdc186
|
si:ch211-225b10.4 |
| chr21_+_45502773 | 0.15 |
ENSDART00000160059
ENSDART00000165704 |
si:dkey-223p19.2
|
si:dkey-223p19.2 |
| chr15_+_21262917 | 0.15 |
ENSDART00000101000
|
gkup
|
glucuronokinase with putative uridyl pyrophosphorylase |
| chr20_+_25879826 | 0.15 |
ENSDART00000018519
|
zgc:153896
|
zgc:153896 |
| chr5_-_25733745 | 0.15 |
ENSDART00000051566
|
zgc:101016
|
zgc:101016 |
| chr20_-_19511700 | 0.14 |
ENSDART00000040191
|
snx17
|
sorting nexin 17 |
| chr6_-_7776612 | 0.14 |
ENSDART00000190269
|
myh9a
|
myosin, heavy chain 9a, non-muscle |
| chr10_-_32494304 | 0.14 |
ENSDART00000028161
|
uvrag
|
UV radiation resistance associated gene |
| chr19_-_19379084 | 0.14 |
ENSDART00000165206
|
smarcc1b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1b |
| chr25_+_28282274 | 0.14 |
ENSDART00000164502
|
aass
|
aminoadipate-semialdehyde synthase |
| chr21_+_1647990 | 0.14 |
ENSDART00000148540
|
fech
|
ferrochelatase |
| chr2_-_38206034 | 0.14 |
ENSDART00000144518
ENSDART00000137395 |
acin1a
|
apoptotic chromatin condensation inducer 1a |
| chr16_-_17586883 | 0.14 |
ENSDART00000017142
|
m6pr
|
mannose-6-phosphate receptor (cation dependent) |
| chr18_-_39188664 | 0.14 |
ENSDART00000162983
|
mapk6
|
mitogen-activated protein kinase 6 |
| chr12_+_32199651 | 0.13 |
ENSDART00000153027
|
si:ch211-277e21.1
|
si:ch211-277e21.1 |
| chr20_-_43663494 | 0.13 |
ENSDART00000144564
|
BX470188.1
|
|
| chr4_-_15103646 | 0.13 |
ENSDART00000138183
ENSDART00000181044 |
nrf1
|
nuclear respiratory factor 1 |
| chr23_-_32162810 | 0.12 |
ENSDART00000155905
|
nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr17_-_21162821 | 0.12 |
ENSDART00000157283
|
abhd12
|
abhydrolase domain containing 12 |
| chr11_+_24820542 | 0.12 |
ENSDART00000135443
|
kdm5ba
|
lysine (K)-specific demethylase 5Ba |
| chr2_-_32384683 | 0.12 |
ENSDART00000182942
ENSDART00000141757 |
ubtfl
|
upstream binding transcription factor, like |
| chr16_+_42471455 | 0.12 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr12_-_11560794 | 0.11 |
ENSDART00000149098
ENSDART00000169975 |
plekha1b
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1b |
| chr23_+_28809002 | 0.11 |
ENSDART00000134121
ENSDART00000183661 |
pex14
|
peroxisomal biogenesis factor 14 |
| chr14_-_2933185 | 0.11 |
ENSDART00000161677
ENSDART00000162446 ENSDART00000109378 |
si:dkey-201i24.6
|
si:dkey-201i24.6 |
| chr6_-_39649504 | 0.11 |
ENSDART00000179960
ENSDART00000190951 |
larp4ab
|
La ribonucleoprotein domain family, member 4Ab |
| chr12_+_30367079 | 0.11 |
ENSDART00000190112
|
ccdc186
|
si:ch211-225b10.4 |
| chr8_-_46894362 | 0.11 |
ENSDART00000111124
|
acot7
|
acyl-CoA thioesterase 7 |
| chr12_+_30723778 | 0.11 |
ENSDART00000153291
|
slc35f3a
|
solute carrier family 35, member F3a |
| chr20_+_31076488 | 0.11 |
ENSDART00000136255
ENSDART00000008840 |
otofa
|
otoferlin a |
| chr7_-_31321027 | 0.11 |
ENSDART00000186878
|
CR356242.1
|
|
| chr10_-_33297864 | 0.10 |
ENSDART00000163360
|
PRDM15
|
PR/SET domain 15 |
| chr22_+_14051894 | 0.10 |
ENSDART00000142548
|
aox6
|
aldehyde oxidase 6 |
| chr2_-_38363017 | 0.10 |
ENSDART00000088026
|
prmt5
|
protein arginine methyltransferase 5 |
| chr3_+_32832538 | 0.10 |
ENSDART00000139410
|
cd2bp2
|
CD2 (cytoplasmic tail) binding protein 2 |
| chr12_+_47698356 | 0.10 |
ENSDART00000112010
|
lzts2b
|
leucine zipper, putative tumor suppressor 2b |
| chr9_+_50001746 | 0.10 |
ENSDART00000058892
|
slc38a11
|
solute carrier family 38, member 11 |
| chr9_+_7732714 | 0.10 |
ENSDART00000145853
|
si:ch1073-349o24.2
|
si:ch1073-349o24.2 |
| chr8_-_8446668 | 0.10 |
ENSDART00000132700
|
cdk16
|
cyclin-dependent kinase 16 |
| chr18_-_43884044 | 0.10 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr3_+_45368973 | 0.10 |
ENSDART00000187282
|
ube2ia
|
ubiquitin-conjugating enzyme E2Ia |
| chr18_+_15644559 | 0.10 |
ENSDART00000061794
|
nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr23_+_16638639 | 0.09 |
ENSDART00000143545
|
snphb
|
syntaphilin b |
| chr7_+_48667081 | 0.09 |
ENSDART00000083473
|
trpm5
|
transient receptor potential cation channel, subfamily M, member 5 |
| chr15_-_17138640 | 0.09 |
ENSDART00000080777
|
mrpl28
|
mitochondrial ribosomal protein L28 |
| chr10_-_8060573 | 0.09 |
ENSDART00000147104
ENSDART00000099030 |
si:ch211-251f6.6
|
si:ch211-251f6.6 |
| chr10_+_7593185 | 0.09 |
ENSDART00000162617
ENSDART00000162590 ENSDART00000171744 |
ppp2cb
|
protein phosphatase 2, catalytic subunit, beta isozyme |
| chr9_+_22485343 | 0.09 |
ENSDART00000146028
|
dgkg
|
diacylglycerol kinase, gamma |
| chr2_+_37227011 | 0.09 |
ENSDART00000126587
ENSDART00000084958 |
samd7
|
sterile alpha motif domain containing 7 |
| chr11_+_12052791 | 0.09 |
ENSDART00000158479
|
si:ch211-156l18.8
|
si:ch211-156l18.8 |
| chr18_+_19456648 | 0.08 |
ENSDART00000079695
|
zwilch
|
zwilch kinetochore protein |
| chr22_+_30543437 | 0.08 |
ENSDART00000137983
|
si:dkey-103k4.1
|
si:dkey-103k4.1 |
| chr6_+_14301214 | 0.08 |
ENSDART00000129491
|
tmem182b
|
transmembrane protein 182b |
| chr20_+_46492175 | 0.08 |
ENSDART00000060695
|
zc3h14
|
zinc finger CCCH-type containing 14 |
| chr6_-_40029423 | 0.08 |
ENSDART00000103230
|
pfkfb4b
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4b |
| chr19_-_5103141 | 0.08 |
ENSDART00000150952
|
tpi1a
|
triosephosphate isomerase 1a |
| chr7_+_19552381 | 0.08 |
ENSDART00000169060
|
si:ch211-212k18.5
|
si:ch211-212k18.5 |
| chr19_+_15440841 | 0.08 |
ENSDART00000182329
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr11_-_22303678 | 0.08 |
ENSDART00000159527
ENSDART00000159681 |
tfeb
|
transcription factor EB |
| chr18_+_8320165 | 0.08 |
ENSDART00000092053
|
chkb
|
choline kinase beta |
| chr19_-_44065581 | 0.08 |
ENSDART00000006338
|
mterf3
|
mitochondrial transcription termination factor 3 |
| chr4_+_306036 | 0.08 |
ENSDART00000103659
|
msgn1
|
mesogenin 1 |
| chr15_+_857148 | 0.08 |
ENSDART00000156949
|
si:dkey-7i4.13
|
si:dkey-7i4.13 |
| chr5_+_44805269 | 0.07 |
ENSDART00000136965
|
ctsla
|
cathepsin La |
| chr11_-_19775182 | 0.07 |
ENSDART00000037894
|
namptb
|
nicotinamide phosphoribosyltransferase b |
| chr3_-_26806032 | 0.07 |
ENSDART00000143710
|
pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr13_-_8692860 | 0.07 |
ENSDART00000058107
|
mcfd2
|
multiple coagulation factor deficiency 2 |
| chr12_+_45238292 | 0.07 |
ENSDART00000057983
|
mrpl38
|
mitochondrial ribosomal protein L38 |
| chr5_+_25733774 | 0.07 |
ENSDART00000137088
ENSDART00000098467 |
abhd17b
|
abhydrolase domain containing 17B |
| chr5_+_66433287 | 0.07 |
ENSDART00000170757
|
kntc1
|
kinetochore associated 1 |
| chr25_-_27564205 | 0.06 |
ENSDART00000157319
|
hyal4
|
hyaluronoglucosaminidase 4 |
| chr7_+_23292133 | 0.06 |
ENSDART00000134489
|
htr2cl1
|
5-hydroxytryptamine (serotonin) receptor 2C, G protein-coupled-like 1 |
| chr22_+_16308806 | 0.06 |
ENSDART00000162685
|
lrrc39
|
leucine rich repeat containing 39 |
| chr12_-_33972798 | 0.06 |
ENSDART00000105545
|
arl3
|
ADP-ribosylation factor-like 3 |
| chr25_-_6389713 | 0.06 |
ENSDART00000083539
|
sin3aa
|
SIN3 transcription regulator family member Aa |
| chr24_+_5912635 | 0.06 |
ENSDART00000153736
|
pimr63
|
Pim proto-oncogene, serine/threonine kinase, related 63 |
| chr9_+_22364997 | 0.06 |
ENSDART00000188054
ENSDART00000046116 |
crygs3
|
crystallin, gamma S3 |
| chr19_+_30884706 | 0.06 |
ENSDART00000052126
|
yars
|
tyrosyl-tRNA synthetase |
| chr21_-_25722834 | 0.06 |
ENSDART00000101208
|
abhd11
|
abhydrolase domain containing 11 |
| chr2_-_28671139 | 0.06 |
ENSDART00000165272
ENSDART00000164657 |
dhcr7
|
7-dehydrocholesterol reductase |
| chr14_-_22113600 | 0.06 |
ENSDART00000113752
|
si:dkey-6i22.5
|
si:dkey-6i22.5 |
| chr8_-_16406440 | 0.05 |
ENSDART00000034004
|
faf1
|
Fas (TNFRSF6) associated factor 1 |
| chr10_+_40660772 | 0.05 |
ENSDART00000148007
|
taar19l
|
trace amine associated receptor 19l |
| chr20_-_7128612 | 0.05 |
ENSDART00000146755
ENSDART00000036871 |
si:ch211-121a2.4
|
si:ch211-121a2.4 |
| chr22_-_7129631 | 0.05 |
ENSDART00000171359
|
asic1b
|
acid-sensing (proton-gated) ion channel 1b |
| chr15_+_42933236 | 0.05 |
ENSDART00000167763
|
slc8a2b
|
solute carrier family 8 (sodium/calcium exchanger), member 2b |
| chr6_+_3280939 | 0.05 |
ENSDART00000151359
|
kdm4aa
|
lysine (K)-specific demethylase 4A, genome duplicate a |
| chr22_+_10440991 | 0.05 |
ENSDART00000064805
|
cenpp
|
centromere protein P |
| chr11_+_33818179 | 0.05 |
ENSDART00000109418
|
spoplb
|
speckle-type POZ protein-like b |
| chr5_-_28109416 | 0.05 |
ENSDART00000042002
|
si:ch211-48m9.1
|
si:ch211-48m9.1 |
| chr13_-_37465524 | 0.04 |
ENSDART00000100324
ENSDART00000147336 |
ppp2r5eb
|
protein phosphatase 2, regulatory subunit B', epsilon isoform b |
| chr2_-_51303937 | 0.04 |
ENSDART00000164111
|
si:ch211-215e19.8
|
si:ch211-215e19.8 |
| chr20_-_40755614 | 0.04 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr25_-_27722614 | 0.03 |
ENSDART00000190154
|
zgc:153935
|
zgc:153935 |
| chr7_-_44957503 | 0.03 |
ENSDART00000165077
|
cdh16
|
cadherin 16, KSP-cadherin |
| chr10_+_11282883 | 0.03 |
ENSDART00000135355
|
si:ch211-126i22.5
|
si:ch211-126i22.5 |
| chr2_-_22927958 | 0.03 |
ENSDART00000141621
|
myo7bb
|
myosin VIIBb |
| chr5_+_19343880 | 0.03 |
ENSDART00000148130
|
acacb
|
acetyl-CoA carboxylase beta |
| chr6_-_51386656 | 0.03 |
ENSDART00000154732
ENSDART00000177990 ENSDART00000184928 ENSDART00000180197 |
ptprt
|
protein tyrosine phosphatase, receptor type, t |
| chr13_-_25819825 | 0.03 |
ENSDART00000077612
|
rel
|
v-rel avian reticuloendotheliosis viral oncogene homolog |
| chr3_-_16719244 | 0.03 |
ENSDART00000055859
|
pold1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr3_-_36210344 | 0.03 |
ENSDART00000025326
|
csnk1da
|
casein kinase 1, delta a |
| chr25_-_4313699 | 0.03 |
ENSDART00000154038
|
syt7a
|
synaptotagmin VIIa |
| chr1_-_44812014 | 0.03 |
ENSDART00000139044
|
si:dkey-9i23.8
|
si:dkey-9i23.8 |
| chr5_+_66063807 | 0.03 |
ENSDART00000122405
|
jak2b
|
Janus kinase 2b |
| chr13_-_36034582 | 0.03 |
ENSDART00000133565
|
si:dkey-157l19.2
|
si:dkey-157l19.2 |
| chr11_-_25539323 | 0.02 |
ENSDART00000155785
|
si:dkey-245f22.3
|
si:dkey-245f22.3 |
| chr3_+_13624815 | 0.02 |
ENSDART00000161451
|
pglyrp6
|
peptidoglycan recognition protein 6 |
| chr17_-_37395460 | 0.02 |
ENSDART00000148160
ENSDART00000075975 |
crip1
|
cysteine-rich protein 1 |
| chr14_+_25817628 | 0.02 |
ENSDART00000047680
|
glra1
|
glycine receptor, alpha 1 |
| chr6_+_41191482 | 0.02 |
ENSDART00000000877
|
opn1mw3
|
opsin 1 (cone pigments), medium-wave-sensitive, 3 |
| chr17_+_7513673 | 0.02 |
ENSDART00000156674
|
klhl10b.1
|
kelch-like family member 10b, tandem duplicate 1 |
| chr17_+_30369396 | 0.02 |
ENSDART00000076611
|
greb1
|
growth regulation by estrogen in breast cancer 1 |
| chr10_+_40568735 | 0.02 |
ENSDART00000136468
ENSDART00000182841 |
taar18i
|
trace amine associated receptor 18i |
| chr1_+_10318089 | 0.01 |
ENSDART00000029774
|
pip4p1b
|
phosphatidylinositol-4,5-bisphosphate 4-phosphatase 1b |
| chr5_+_20147830 | 0.01 |
ENSDART00000098727
|
svopa
|
SV2 related protein a |
| chr18_-_33979422 | 0.01 |
ENSDART00000136535
ENSDART00000167698 |
si:ch211-203b20.7
|
si:ch211-203b20.7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of mnx1+mnx2a+mnx2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0039015 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.1 | 0.4 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.1 | 0.2 | GO:0097065 | anterior head development(GO:0097065) |
| 0.1 | 0.5 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.1 | 0.2 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.1 | 0.6 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0009085 | lysine biosynthetic process(GO:0009085) lysine biosynthetic process via aminoadipic acid(GO:0019878) L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.0 | 0.1 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.0 | 0.5 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.3 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.6 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:0072103 | renal system vasculature morphogenesis(GO:0061438) kidney vasculature morphogenesis(GO:0061439) glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.0 | 0.3 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.1 | GO:0031441 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.3 | GO:0030816 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.0 | 0.1 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 0.0 | 0.1 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.0 | 0.1 | GO:0048340 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) |
| 0.0 | 0.1 | GO:0046462 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.4 | GO:0051445 | regulation of meiotic cell cycle(GO:0051445) |
| 0.0 | 0.1 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.0 | 0.5 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.0 | 0.3 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.1 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 0.2 | GO:0015886 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.0 | 0.3 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.0 | 0.3 | GO:0035803 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.1 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.0 | 0.2 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.2 | GO:0090308 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.1 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.1 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.3 | GO:0008345 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.1 | GO:0022615 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.0 | 0.1 | GO:0050909 | sensory perception of taste(GO:0050909) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.0 | 0.2 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.0 | 0.5 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.3 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.0 | 0.1 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.0 | 1.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 1.0 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0098833 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.0 | 0.1 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.0 | 0.4 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.1 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0009374 | biotin binding(GO:0009374) |
| 0.1 | 0.9 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 0.5 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 0.2 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.1 | 0.2 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 0.2 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.3 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.0 | 0.5 | GO:0031729 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.0 | 0.6 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.1 | GO:0016623 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.1 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 0.3 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.7 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.2 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.1 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.0 | 0.7 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.0 | 0.5 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.3 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.4 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.1 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.2 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.3 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.3 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.1 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.1 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.8 | GO:0003724 | RNA helicase activity(GO:0003724) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.5 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |