Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
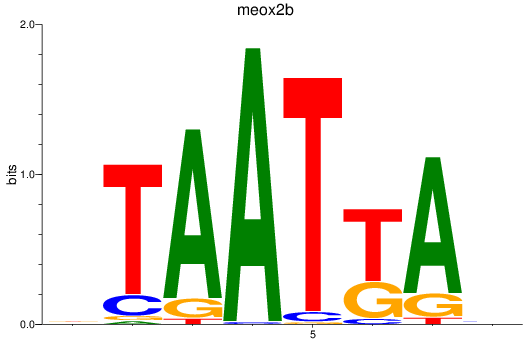
Results for meox2b
Z-value: 0.72
Transcription factors associated with meox2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
meox2b
|
ENSDARG00000061818 | mesenchyme homeobox 2b |
|
meox2b
|
ENSDARG00000116206 | mesenchyme homeobox 2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| meox2b | dr11_v1_chr19_+_31183495_31183495 | 0.38 | 3.1e-01 | Click! |
Activity profile of meox2b motif
Sorted Z-values of meox2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_+_14507567 | 1.56 |
ENSDART00000015681
|
dbx1b
|
developing brain homeobox 1b |
| chr23_-_23401305 | 1.39 |
ENSDART00000078936
|
her9
|
hairy-related 9 |
| chr16_+_23431189 | 1.38 |
ENSDART00000004679
|
icn
|
ictacalcin |
| chr16_+_23913943 | 1.37 |
ENSDART00000175404
ENSDART00000129525 |
apoa4b.1
|
apolipoprotein A-IV b, tandem duplicate 1 |
| chr3_+_30922947 | 1.34 |
ENSDART00000184060
|
cldni
|
claudin i |
| chr8_+_21353878 | 1.00 |
ENSDART00000056420
|
alas2
|
aminolevulinate, delta-, synthase 2 |
| chr17_+_15433671 | 0.99 |
ENSDART00000149568
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr25_+_31276842 | 0.98 |
ENSDART00000187238
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr10_+_39084354 | 0.97 |
ENSDART00000158245
|
si:ch73-1a9.3
|
si:ch73-1a9.3 |
| chr15_+_46356879 | 0.95 |
ENSDART00000154388
|
wu:fb18f06
|
wu:fb18f06 |
| chr20_+_2281933 | 0.92 |
ENSDART00000137579
|
si:ch73-18b11.2
|
si:ch73-18b11.2 |
| chr17_+_15433518 | 0.91 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr12_-_35830625 | 0.88 |
ENSDART00000180028
|
CU459056.1
|
|
| chr3_-_25814097 | 0.84 |
ENSDART00000169706
|
ntn1b
|
netrin 1b |
| chr25_-_31423493 | 0.83 |
ENSDART00000027661
|
myod1
|
myogenic differentiation 1 |
| chr1_+_52392511 | 0.83 |
ENSDART00000144025
|
si:ch211-217k17.8
|
si:ch211-217k17.8 |
| chr12_+_27117609 | 0.80 |
ENSDART00000076154
|
hoxb8b
|
homeobox B8b |
| chr14_+_45406299 | 0.79 |
ENSDART00000173142
ENSDART00000112377 |
map1lc3cl
|
microtubule-associated protein 1 light chain 3 gamma, like |
| chr13_+_25449681 | 0.79 |
ENSDART00000101328
|
atoh7
|
atonal bHLH transcription factor 7 |
| chr15_-_21877726 | 0.78 |
ENSDART00000127819
ENSDART00000145646 ENSDART00000100897 ENSDART00000144739 |
zgc:162608
|
zgc:162608 |
| chr7_-_25895189 | 0.78 |
ENSDART00000173599
ENSDART00000079235 ENSDART00000173786 ENSDART00000173602 ENSDART00000079245 ENSDART00000187568 ENSDART00000173505 |
cd99l2
|
CD99 molecule-like 2 |
| chr6_+_15268685 | 0.78 |
ENSDART00000128090
ENSDART00000154417 |
ecrg4b
|
esophageal cancer related gene 4b |
| chr3_+_46635527 | 0.77 |
ENSDART00000153971
|
si:dkey-248g21.1
|
si:dkey-248g21.1 |
| chr24_-_21923930 | 0.74 |
ENSDART00000131944
|
tagln3b
|
transgelin 3b |
| chr7_-_2039060 | 0.74 |
ENSDART00000173879
|
si:cabz01007794.1
|
si:cabz01007794.1 |
| chr2_+_27010439 | 0.72 |
ENSDART00000030547
|
cdh7a
|
cadherin 7a |
| chr16_+_5774977 | 0.72 |
ENSDART00000134202
|
ccka
|
cholecystokinin a |
| chr19_-_42607451 | 0.72 |
ENSDART00000004392
|
fkbp9
|
FK506 binding protein 9 |
| chr24_+_17269849 | 0.71 |
ENSDART00000017605
|
spag6
|
sperm associated antigen 6 |
| chr3_+_28953274 | 0.70 |
ENSDART00000133528
ENSDART00000103602 |
lgals2a
|
lectin, galactoside-binding, soluble, 2a |
| chr4_-_2545310 | 0.70 |
ENSDART00000150619
ENSDART00000140760 |
e2f7
|
E2F transcription factor 7 |
| chr16_-_24044664 | 0.70 |
ENSDART00000136982
|
bcam
|
basal cell adhesion molecule (Lutheran blood group) |
| chr12_+_20352400 | 0.70 |
ENSDART00000066383
|
hbae5
|
hemoglobin, alpha embryonic 5 |
| chr11_-_45138857 | 0.69 |
ENSDART00000166501
|
cant1b
|
calcium activated nucleotidase 1b |
| chr15_+_23799461 | 0.69 |
ENSDART00000154885
|
si:ch211-167j9.4
|
si:ch211-167j9.4 |
| chr10_-_24391716 | 0.68 |
ENSDART00000141332
ENSDART00000100772 |
slc43a2b
|
solute carrier family 43 (amino acid system L transporter), member 2b |
| chr17_-_14876758 | 0.68 |
ENSDART00000155857
|
nid2a
|
nidogen 2a (osteonidogen) |
| chr11_-_1509773 | 0.68 |
ENSDART00000050762
|
phactr3b
|
phosphatase and actin regulator 3b |
| chr4_+_12615836 | 0.68 |
ENSDART00000003583
|
lmo3
|
LIM domain only 3 |
| chr9_-_33107237 | 0.67 |
ENSDART00000013918
|
casq2
|
calsequestrin 2 |
| chr22_-_10459880 | 0.67 |
ENSDART00000064801
|
ogn
|
osteoglycin |
| chr1_-_45049603 | 0.65 |
ENSDART00000023336
|
rps6
|
ribosomal protein S6 |
| chr11_+_3254524 | 0.64 |
ENSDART00000159459
|
pmela
|
premelanosome protein a |
| chr16_+_23961276 | 0.64 |
ENSDART00000192754
|
apoeb
|
apolipoprotein Eb |
| chr10_+_42358426 | 0.64 |
ENSDART00000025691
|
dbnla
|
drebrin-like a |
| chr24_+_5237753 | 0.63 |
ENSDART00000106488
ENSDART00000005901 |
plod2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 |
| chr18_-_33979693 | 0.63 |
ENSDART00000021215
|
si:ch211-203b20.7
|
si:ch211-203b20.7 |
| chr21_-_37973819 | 0.62 |
ENSDART00000133405
|
ripply1
|
ripply transcriptional repressor 1 |
| chr5_-_41307550 | 0.62 |
ENSDART00000143446
|
npr3
|
natriuretic peptide receptor 3 |
| chr19_+_12915498 | 0.62 |
ENSDART00000132892
|
cthrc1a
|
collagen triple helix repeat containing 1a |
| chr13_+_30903816 | 0.62 |
ENSDART00000191727
|
ercc6
|
excision repair cross-complementation group 6 |
| chr25_+_29160102 | 0.62 |
ENSDART00000162854
|
pkmb
|
pyruvate kinase M1/2b |
| chr7_+_20503344 | 0.61 |
ENSDART00000157699
|
si:dkey-19b23.12
|
si:dkey-19b23.12 |
| chr16_-_7443388 | 0.61 |
ENSDART00000017445
|
prdm1a
|
PR domain containing 1a, with ZNF domain |
| chr22_-_15593824 | 0.61 |
ENSDART00000123125
|
tpm4a
|
tropomyosin 4a |
| chr22_+_16535575 | 0.61 |
ENSDART00000083063
|
tal1
|
T-cell acute lymphocytic leukemia 1 |
| chr4_+_14900042 | 0.59 |
ENSDART00000018261
|
akr1b1
|
aldo-keto reductase family 1, member B1 (aldose reductase) |
| chr21_+_28445052 | 0.59 |
ENSDART00000077871
|
pygma
|
phosphorylase, glycogen, muscle A |
| chr23_+_42482137 | 0.59 |
ENSDART00000160199
|
cyp2aa3
|
cytochrome P450, family 2, subfamily AA, polypeptide 3 |
| chr14_-_4273396 | 0.59 |
ENSDART00000127318
|
frmpd1b
|
FERM and PDZ domain containing 1b |
| chr11_+_44135351 | 0.58 |
ENSDART00000182914
|
FO704721.1
|
|
| chr18_+_29402623 | 0.56 |
ENSDART00000014703
|
mafa
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog a (paralog a) |
| chr17_+_23298928 | 0.56 |
ENSDART00000153652
|
zgc:165461
|
zgc:165461 |
| chr11_+_14199802 | 0.55 |
ENSDART00000102520
ENSDART00000133172 |
palm1a
|
paralemmin 1a |
| chr9_-_21918963 | 0.55 |
ENSDART00000090782
|
lmo7a
|
LIM domain 7a |
| chr24_-_39610585 | 0.54 |
ENSDART00000066506
|
cox6b1
|
cytochrome c oxidase subunit VIb polypeptide 1 |
| chr6_-_54826061 | 0.54 |
ENSDART00000149982
|
tnni1b
|
troponin I type 1b (skeletal, slow) |
| chr14_-_3381303 | 0.54 |
ENSDART00000171601
|
im:7150988
|
im:7150988 |
| chr2_-_23172708 | 0.54 |
ENSDART00000041365
|
prrx1a
|
paired related homeobox 1a |
| chr16_+_1353894 | 0.54 |
ENSDART00000148426
|
celf3b
|
cugbp, Elav-like family member 3b |
| chr7_-_48667056 | 0.54 |
ENSDART00000006378
|
cdkn1ca
|
cyclin-dependent kinase inhibitor 1Ca |
| chr23_+_20110086 | 0.54 |
ENSDART00000054664
|
tnnc1b
|
troponin C type 1b (slow) |
| chr12_-_5728755 | 0.53 |
ENSDART00000105887
|
dlx4b
|
distal-less homeobox 4b |
| chr2_-_10703621 | 0.53 |
ENSDART00000005944
|
rpl5a
|
ribosomal protein L5a |
| chr16_+_23984179 | 0.53 |
ENSDART00000175879
|
apoc2
|
apolipoprotein C-II |
| chr16_-_45235947 | 0.53 |
ENSDART00000164436
|
si:dkey-33i11.4
|
si:dkey-33i11.4 |
| chr11_+_8565323 | 0.52 |
ENSDART00000081607
ENSDART00000141612 |
tbl1xr1a
|
transducin (beta)-like 1 X-linked receptor 1a |
| chr13_+_27232848 | 0.52 |
ENSDART00000138043
|
rin2
|
Ras and Rab interactor 2 |
| chr1_-_58036509 | 0.52 |
ENSDART00000081122
|
COLGALT1
|
si:ch211-114l13.7 |
| chr15_+_19652807 | 0.51 |
ENSDART00000134321
ENSDART00000054426 |
lim2.3
|
lens intrinsic membrane protein 2.3 |
| chr13_+_33282095 | 0.51 |
ENSDART00000135200
|
ccdc28b
|
coiled-coil domain containing 28B |
| chr24_-_6078222 | 0.51 |
ENSDART00000146830
|
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr12_-_10409961 | 0.50 |
ENSDART00000149521
ENSDART00000052001 |
eef2k
|
eukaryotic elongation factor 2 kinase |
| chr15_-_28596507 | 0.50 |
ENSDART00000156800
|
si:ch211-225b7.5
|
si:ch211-225b7.5 |
| chr8_-_19216657 | 0.50 |
ENSDART00000135096
ENSDART00000135869 ENSDART00000145951 |
si:ch73-222f22.2
si:ch73-222f22.2
|
si:ch73-222f22.2 si:ch73-222f22.2 |
| chr9_+_31282161 | 0.50 |
ENSDART00000010774
|
zic2a
|
zic family member 2 (odd-paired homolog, Drosophila), a |
| chr23_+_43489182 | 0.50 |
ENSDART00000162062
|
znf341
|
zinc finger protein 341 |
| chr19_-_41069573 | 0.50 |
ENSDART00000111982
ENSDART00000193142 |
sgce
|
sarcoglycan, epsilon |
| chr9_-_18911608 | 0.49 |
ENSDART00000138785
|
si:dkey-239h2.3
|
si:dkey-239h2.3 |
| chr1_-_54063520 | 0.49 |
ENSDART00000171722
|
smdt1b
|
single-pass membrane protein with aspartate-rich tail 1b |
| chr4_+_9669717 | 0.49 |
ENSDART00000004604
|
si:dkey-153k10.9
|
si:dkey-153k10.9 |
| chr18_-_19456269 | 0.49 |
ENSDART00000060363
|
rpl4
|
ribosomal protein L4 |
| chr14_-_21064199 | 0.49 |
ENSDART00000172099
|
si:dkey-74k8.3
|
si:dkey-74k8.3 |
| chr11_-_30158191 | 0.49 |
ENSDART00000155278
ENSDART00000156121 |
scml2
|
Scm polycomb group protein like 2 |
| chr1_-_45633955 | 0.49 |
ENSDART00000044057
|
sept3
|
septin 3 |
| chr19_+_43297546 | 0.48 |
ENSDART00000168002
|
laptm5
|
lysosomal protein transmembrane 5 |
| chr20_+_539852 | 0.48 |
ENSDART00000185994
|
dse
|
dermatan sulfate epimerase |
| chr21_+_28958471 | 0.48 |
ENSDART00000144331
ENSDART00000005929 |
ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr11_-_41853874 | 0.48 |
ENSDART00000002556
|
mrto4
|
MRT4 homolog, ribosome maturation factor |
| chr17_-_25649079 | 0.48 |
ENSDART00000130955
|
ppp1cb
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr21_-_5205617 | 0.48 |
ENSDART00000145554
ENSDART00000045284 |
rpl37
|
ribosomal protein L37 |
| chr7_+_34290051 | 0.48 |
ENSDART00000123498
|
fibinb
|
fin bud initiation factor b |
| chr7_-_24699985 | 0.48 |
ENSDART00000052802
|
calb2b
|
calbindin 2b |
| chr23_+_19813677 | 0.47 |
ENSDART00000139192
ENSDART00000142308 |
emd
|
emerin (Emery-Dreifuss muscular dystrophy) |
| chr24_+_9744012 | 0.47 |
ENSDART00000129656
|
tmem108
|
transmembrane protein 108 |
| chr1_+_52929185 | 0.47 |
ENSDART00000147683
|
inpp4b
|
inositol polyphosphate-4-phosphatase type II B |
| chr21_+_27416284 | 0.47 |
ENSDART00000077593
ENSDART00000108763 |
cfb
|
complement factor B |
| chr6_+_47424266 | 0.47 |
ENSDART00000171087
|
si:ch211-286o17.1
|
si:ch211-286o17.1 |
| chr3_-_23643751 | 0.46 |
ENSDART00000078425
ENSDART00000140264 |
eve1
|
even-skipped-like1 |
| chr4_+_12612145 | 0.46 |
ENSDART00000181201
|
lmo3
|
LIM domain only 3 |
| chr4_-_16341801 | 0.46 |
ENSDART00000140190
|
kera
|
keratocan |
| chr7_-_54679595 | 0.46 |
ENSDART00000165320
|
ccnd1
|
cyclin D1 |
| chr16_+_33655890 | 0.46 |
ENSDART00000143757
|
fhl3a
|
four and a half LIM domains 3a |
| chr3_+_18398876 | 0.46 |
ENSDART00000141100
ENSDART00000138107 |
rps2
|
ribosomal protein S2 |
| chr23_+_36095260 | 0.46 |
ENSDART00000127384
|
hoxc9a
|
homeobox C9a |
| chr14_+_3287740 | 0.46 |
ENSDART00000186290
|
cdx1a
|
caudal type homeobox 1a |
| chr15_-_47865063 | 0.45 |
ENSDART00000151600
|
h3f3b.1
|
H3 histone, family 3B.1 |
| chr1_-_40911332 | 0.45 |
ENSDART00000027463
|
hmx4
|
H6 family homeobox 4 |
| chr20_+_6659770 | 0.45 |
ENSDART00000192135
|
tns3.2
|
tensin 3, tandem duplicate 2 |
| chr16_+_50972803 | 0.45 |
ENSDART00000178189
|
si:dkeyp-97a10.2
|
si:dkeyp-97a10.2 |
| chr9_+_25776971 | 0.45 |
ENSDART00000146011
|
zeb2a
|
zinc finger E-box binding homeobox 2a |
| chr15_-_22074315 | 0.45 |
ENSDART00000149830
|
drd2a
|
dopamine receptor D2a |
| chr20_-_40370736 | 0.45 |
ENSDART00000041229
|
fabp7b
|
fatty acid binding protein 7, brain, b |
| chr17_+_21295132 | 0.45 |
ENSDART00000103845
|
eno4
|
enolase family member 4 |
| chr1_+_34696503 | 0.44 |
ENSDART00000186106
|
CR339054.2
|
|
| chr16_+_46111849 | 0.44 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr13_-_31296358 | 0.44 |
ENSDART00000030946
|
prdm8
|
PR domain containing 8 |
| chr16_-_42933332 | 0.44 |
ENSDART00000057305
|
thbs3a
|
thrombospondin 3a |
| chr2_-_33993533 | 0.44 |
ENSDART00000140910
ENSDART00000077304 |
ptch2
|
patched 2 |
| chr11_-_6048490 | 0.44 |
ENSDART00000066164
|
plvapb
|
plasmalemma vesicle associated protein b |
| chr3_-_30685401 | 0.44 |
ENSDART00000151097
|
si:ch211-51c14.1
|
si:ch211-51c14.1 |
| chr24_-_25691020 | 0.44 |
ENSDART00000015391
|
chrnd
|
cholinergic receptor, nicotinic, delta (muscle) |
| chr1_+_12767318 | 0.44 |
ENSDART00000162652
|
pcdh10a
|
protocadherin 10a |
| chr13_-_31470439 | 0.43 |
ENSDART00000076574
|
rtn1a
|
reticulon 1a |
| chr11_+_2198831 | 0.43 |
ENSDART00000160515
|
hoxc6b
|
homeobox C6b |
| chr2_-_30668580 | 0.43 |
ENSDART00000087270
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr10_-_3295197 | 0.43 |
ENSDART00000109131
|
slc25a1b
|
slc25a1 solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1b |
| chr20_-_45812144 | 0.43 |
ENSDART00000147897
ENSDART00000147637 |
fermt1
|
fermitin family member 1 |
| chr9_-_12424791 | 0.42 |
ENSDART00000135447
ENSDART00000088199 |
zgc:162707
|
zgc:162707 |
| chr7_-_58130703 | 0.42 |
ENSDART00000172082
|
ank2b
|
ankyrin 2b, neuronal |
| chr5_+_23242370 | 0.42 |
ENSDART00000051532
|
agtr2
|
angiotensin II receptor, type 2 |
| chr23_+_11285662 | 0.42 |
ENSDART00000111028
|
chl1a
|
cell adhesion molecule L1-like a |
| chr16_+_16969060 | 0.42 |
ENSDART00000182819
ENSDART00000191876 |
si:ch211-120k19.1
rpl18
|
si:ch211-120k19.1 ribosomal protein L18 |
| chr2_-_32356539 | 0.42 |
ENSDART00000169316
|
asap1a
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1a |
| chr23_+_28731379 | 0.42 |
ENSDART00000047378
|
cort
|
cortistatin |
| chr24_+_21621654 | 0.41 |
ENSDART00000002595
|
rpl21
|
ribosomal protein L21 |
| chr20_-_27225876 | 0.41 |
ENSDART00000149204
ENSDART00000149732 |
si:dkey-85n7.7
|
si:dkey-85n7.7 |
| chr21_-_22827548 | 0.41 |
ENSDART00000079161
|
angptl5
|
angiopoietin-like 5 |
| chr21_-_26495700 | 0.41 |
ENSDART00000109379
|
cd248b
|
CD248 molecule, endosialin b |
| chr8_+_28259347 | 0.41 |
ENSDART00000110857
|
fam212b
|
family with sequence similarity 212, member B |
| chr15_+_36309070 | 0.40 |
ENSDART00000157034
|
gmnc
|
geminin coiled-coil domain containing |
| chr22_+_12770877 | 0.40 |
ENSDART00000044683
|
ftcd
|
formimidoyltransferase cyclodeaminase |
| chr20_+_15015557 | 0.40 |
ENSDART00000039345
|
myoc
|
myocilin |
| chr5_+_27897504 | 0.40 |
ENSDART00000130936
|
adam28
|
ADAM metallopeptidase domain 28 |
| chr4_+_43522680 | 0.40 |
ENSDART00000182252
|
si:dkeyp-53e4.4
|
si:dkeyp-53e4.4 |
| chr6_+_52804267 | 0.40 |
ENSDART00000065681
|
matn4
|
matrilin 4 |
| chr16_+_40576679 | 0.40 |
ENSDART00000169412
ENSDART00000193464 |
ccne2
|
cyclin E2 |
| chr25_+_31868268 | 0.39 |
ENSDART00000022325
|
parp16
|
poly (ADP-ribose) polymerase family, member 16 |
| chr24_+_9412450 | 0.39 |
ENSDART00000132724
|
si:ch211-285f17.1
|
si:ch211-285f17.1 |
| chr16_-_28878080 | 0.39 |
ENSDART00000149501
|
si:dkey-239n17.4
|
si:dkey-239n17.4 |
| chr17_+_24687338 | 0.39 |
ENSDART00000135794
|
selenon
|
selenoprotein N |
| chr13_+_35339182 | 0.39 |
ENSDART00000019323
|
jag1b
|
jagged 1b |
| chr5_-_48268049 | 0.39 |
ENSDART00000187454
|
mef2cb
|
myocyte enhancer factor 2cb |
| chr18_+_40355408 | 0.39 |
ENSDART00000167134
|
sema6dl
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D, like |
| chr20_-_7072487 | 0.39 |
ENSDART00000145954
|
si:ch211-121a2.2
|
si:ch211-121a2.2 |
| chr13_+_255067 | 0.38 |
ENSDART00000102505
|
foxg1d
|
forkhead box G1d |
| chr21_-_37194365 | 0.38 |
ENSDART00000100286
|
fgfr4
|
fibroblast growth factor receptor 4 |
| chr5_-_20205075 | 0.38 |
ENSDART00000051611
|
dao.3
|
D-amino-acid oxidase, tandem duplicate 3 |
| chr14_-_17072736 | 0.38 |
ENSDART00000106333
|
phox2bb
|
paired-like homeobox 2bb |
| chr12_+_24952902 | 0.38 |
ENSDART00000189086
ENSDART00000014868 |
calm3a
|
calmodulin 3a (phosphorylase kinase, delta) |
| chr21_-_25604603 | 0.38 |
ENSDART00000133134
ENSDART00000139460 |
efemp2b
|
EGF containing fibulin extracellular matrix protein 2b |
| chr7_-_12968689 | 0.38 |
ENSDART00000173115
ENSDART00000013690 |
rplp2l
|
ribosomal protein, large P2, like |
| chr7_-_28148310 | 0.38 |
ENSDART00000044208
|
lmo1
|
LIM domain only 1 |
| chr21_-_19314618 | 0.37 |
ENSDART00000188744
|
gpat3
|
glycerol-3-phosphate acyltransferase 3 |
| chr21_+_20383837 | 0.37 |
ENSDART00000026430
|
hspb11
|
heat shock protein, alpha-crystallin-related, b11 |
| chr12_+_8168272 | 0.37 |
ENSDART00000054092
|
arid5b
|
AT-rich interaction domain 5B |
| chr3_-_60175470 | 0.37 |
ENSDART00000156597
|
si:ch73-364h19.1
|
si:ch73-364h19.1 |
| chr1_+_54865552 | 0.37 |
ENSDART00000145381
|
si:ch211-196h16.5
|
si:ch211-196h16.5 |
| chr21_+_19062124 | 0.37 |
ENSDART00000134746
|
rpl17
|
ribosomal protein L17 |
| chr24_+_22485710 | 0.36 |
ENSDART00000146058
|
si:dkey-40h20.1
|
si:dkey-40h20.1 |
| chr16_+_31804590 | 0.36 |
ENSDART00000167321
|
wnt4b
|
wingless-type MMTV integration site family, member 4b |
| chr18_+_3037998 | 0.36 |
ENSDART00000185844
ENSDART00000162657 |
rps3
|
ribosomal protein S3 |
| chr13_+_7442023 | 0.36 |
ENSDART00000080975
|
tnfaip2b
|
tumor necrosis factor, alpha-induced protein 2b |
| chr24_-_40860603 | 0.36 |
ENSDART00000188032
|
CU633479.7
|
|
| chr6_-_39270851 | 0.36 |
ENSDART00000148839
|
arhgef25b
|
Rho guanine nucleotide exchange factor (GEF) 25b |
| chr4_+_391297 | 0.36 |
ENSDART00000030215
|
rpl18a
|
ribosomal protein L18a |
| chr24_-_4450238 | 0.36 |
ENSDART00000066835
|
fzd8a
|
frizzled class receptor 8a |
| chr7_+_69019851 | 0.35 |
ENSDART00000162891
|
CABZ01057488.1
|
|
| chr7_-_38792543 | 0.35 |
ENSDART00000157416
|
si:dkey-23n7.10
|
si:dkey-23n7.10 |
| chr15_+_7086327 | 0.35 |
ENSDART00000114560
|
pik3cb
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit beta |
| chr15_-_14212777 | 0.35 |
ENSDART00000165572
|
CR925813.1
|
|
| chr4_+_58576146 | 0.35 |
ENSDART00000164911
|
si:ch211-212k5.4
|
si:ch211-212k5.4 |
| chr18_+_30028135 | 0.35 |
ENSDART00000140825
ENSDART00000145306 ENSDART00000136810 |
si:ch211-220f16.1
|
si:ch211-220f16.1 |
| chr20_+_4060839 | 0.35 |
ENSDART00000178565
|
TRIM67
|
tripartite motif containing 67 |
| chr23_+_31107685 | 0.35 |
ENSDART00000103448
|
tbx18
|
T-box 18 |
| chr3_+_17537352 | 0.34 |
ENSDART00000104549
|
hcrt
|
hypocretin (orexin) neuropeptide precursor |
| chr25_+_35553542 | 0.34 |
ENSDART00000113723
|
spi1a
|
Spi-1 proto-oncogene a |
| chr16_+_16968682 | 0.34 |
ENSDART00000111074
|
si:ch211-120k19.1
|
si:ch211-120k19.1 |
| chr11_+_30244356 | 0.34 |
ENSDART00000036050
ENSDART00000150080 |
rs1a
|
retinoschisin 1a |
| chr14_+_41345175 | 0.34 |
ENSDART00000086104
|
nox1
|
NADPH oxidase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of meox2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.3 | 0.8 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.3 | 0.8 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.2 | 0.6 | GO:0097435 | fibril organization(GO:0097435) |
| 0.2 | 0.6 | GO:0034382 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 0.2 | 0.6 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.2 | 0.6 | GO:0052575 | carbohydrate localization(GO:0052575) carbohydrate storage(GO:0052576) |
| 0.2 | 0.5 | GO:0009912 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.2 | 0.5 | GO:0060898 | eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.2 | 0.7 | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling(GO:0010882) |
| 0.2 | 0.6 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.2 | 0.8 | GO:0034380 | regulation of cholesterol esterification(GO:0010872) positive regulation of cholesterol esterification(GO:0010873) high-density lipoprotein particle assembly(GO:0034380) |
| 0.1 | 0.7 | GO:0042023 | regulation of DNA endoreduplication(GO:0032875) positive regulation of DNA endoreduplication(GO:0032877) DNA endoreduplication(GO:0042023) |
| 0.1 | 0.4 | GO:0015942 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.1 | 0.4 | GO:0070857 | regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.1 | 0.5 | GO:0034434 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.1 | 0.5 | GO:1900271 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 0.1 | 0.5 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.1 | 0.8 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.1 | 0.4 | GO:0006843 | mitochondrial citrate transport(GO:0006843) |
| 0.1 | 0.3 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 0.1 | 0.5 | GO:0071480 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.1 | 0.5 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.1 | 0.4 | GO:0009080 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.1 | 0.5 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.1 | 0.4 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.1 | 0.5 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.1 | 0.6 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 | 0.3 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) retrograde trans-synaptic signaling by nitric oxide(GO:0098924) |
| 0.1 | 0.4 | GO:0060231 | mesenchymal to epithelial transition(GO:0060231) |
| 0.1 | 0.3 | GO:0042755 | eating behavior(GO:0042755) |
| 0.1 | 0.4 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.2 | GO:0098801 | renal system process involved in regulation of blood volume(GO:0001977) renal system process involved in regulation of systemic arterial blood pressure(GO:0003071) regulation of glomerular filtration(GO:0003093) regulation of renal system process(GO:0098801) |
| 0.1 | 0.2 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.1 | 0.4 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.1 | 0.2 | GO:1901890 | positive regulation of cell junction assembly(GO:1901890) |
| 0.1 | 0.5 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.1 | 0.3 | GO:0060467 | negative regulation of fertilization(GO:0060467) prevention of polyspermy(GO:0060468) cortical granule exocytosis(GO:0060471) |
| 0.1 | 0.3 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.1 | 0.3 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.1 | 0.3 | GO:0033363 | secretory granule organization(GO:0033363) platelet dense granule organization(GO:0060155) |
| 0.1 | 0.4 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.1 | 0.2 | GO:0032757 | interleukin-8 production(GO:0032637) regulation of interleukin-8 production(GO:0032677) positive regulation of interleukin-8 production(GO:0032757) |
| 0.1 | 0.2 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.1 | 0.6 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.1 | 0.6 | GO:0061075 | positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) |
| 0.1 | 0.3 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) microglia development(GO:0014005) regulation of cellular amine metabolic process(GO:0033238) |
| 0.1 | 0.4 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.1 | 0.5 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.1 | 0.2 | GO:0044038 | cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.1 | 3.0 | GO:0060048 | cardiac muscle contraction(GO:0060048) |
| 0.1 | 0.5 | GO:0086002 | cardiac muscle cell action potential involved in contraction(GO:0086002) |
| 0.1 | 0.3 | GO:0061337 | cardiac conduction(GO:0061337) |
| 0.1 | 0.2 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.1 | 0.3 | GO:0060055 | larval development(GO:0002164) larval heart development(GO:0007508) angiogenesis involved in wound healing(GO:0060055) |
| 0.1 | 0.5 | GO:0097106 | postsynaptic density organization(GO:0097106) |
| 0.1 | 0.2 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.1 | 0.4 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.1 | 0.4 | GO:0050848 | regulation of calcium-mediated signaling(GO:0050848) |
| 0.1 | 0.3 | GO:0006562 | proline catabolic process(GO:0006562) proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.2 | GO:0089709 | L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.1 | 0.2 | GO:0005986 | sucrose metabolic process(GO:0005985) sucrose biosynthetic process(GO:0005986) disaccharide biosynthetic process(GO:0046351) |
| 0.1 | 0.2 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 | 0.2 | GO:0015893 | folic acid transport(GO:0015884) drug transport(GO:0015893) methotrexate transport(GO:0051958) reduced folate transmembrane transport(GO:0098838) |
| 0.1 | 0.3 | GO:1902514 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.1 | 0.2 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.1 | 0.7 | GO:0090303 | positive regulation of wound healing(GO:0090303) |
| 0.0 | 0.8 | GO:0046501 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.3 | GO:0090497 | mesenchymal cell migration(GO:0090497) |
| 0.0 | 0.2 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.0 | 0.7 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.3 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.2 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.1 | GO:1904088 | regulation of convergent extension involved in axis elongation(GO:1901232) positive regulation of epiboly involved in gastrulation with mouth forming second(GO:1904088) |
| 0.0 | 0.2 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.0 | 0.2 | GO:0046084 | adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.0 | 0.1 | GO:0010660 | negative regulation of muscle cell apoptotic process(GO:0010656) muscle cell apoptotic process(GO:0010657) striated muscle cell apoptotic process(GO:0010658) regulation of muscle cell apoptotic process(GO:0010660) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 0.0 | 0.5 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.0 | 0.3 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.0 | 1.0 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.5 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.0 | 0.1 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.1 | GO:0008344 | adult locomotory behavior(GO:0008344) |
| 0.0 | 0.8 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.5 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.0 | 0.2 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.6 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.1 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.0 | 0.6 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.0 | 0.3 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.7 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.2 | GO:0070294 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.3 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.0 | 0.3 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.0 | 0.3 | GO:0035479 | angioblast cell migration from lateral mesoderm to midline(GO:0035479) |
| 0.0 | 0.7 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.3 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.0 | 0.4 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.1 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.1 | GO:0071387 | cellular response to cortisol stimulus(GO:0071387) |
| 0.0 | 0.2 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.4 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.1 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.4 | GO:0030878 | thyroid gland development(GO:0030878) |
| 0.0 | 0.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.1 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to ciliary membrane(GO:1903441) |
| 0.0 | 0.2 | GO:0043584 | nose development(GO:0043584) |
| 0.0 | 0.6 | GO:0060038 | cardiac muscle cell proliferation(GO:0060038) |
| 0.0 | 0.7 | GO:0060034 | notochord cell differentiation(GO:0060034) |
| 0.0 | 0.4 | GO:0033198 | response to ATP(GO:0033198) |
| 0.0 | 0.7 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.2 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.0 | 0.4 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.0 | 0.5 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.3 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.0 | 0.3 | GO:0060118 | vestibular receptor cell differentiation(GO:0060114) vestibular receptor cell development(GO:0060118) |
| 0.0 | 0.7 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.5 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.3 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.1 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.9 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.3 | GO:0000467 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.0 | 0.2 | GO:0035909 | aorta morphogenesis(GO:0035909) dorsal aorta morphogenesis(GO:0035912) |
| 0.0 | 0.7 | GO:0021983 | pituitary gland development(GO:0021983) |
| 0.0 | 0.4 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.2 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 0.1 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.0 | 0.1 | GO:0061188 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.0 | 1.6 | GO:0021515 | cell differentiation in spinal cord(GO:0021515) |
| 0.0 | 0.3 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.2 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.1 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
| 0.0 | 0.2 | GO:0043092 | amino acid import(GO:0043090) L-amino acid import(GO:0043092) |
| 0.0 | 0.1 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.0 | 0.1 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.2 | GO:0046959 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.0 | 0.4 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.0 | 0.2 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.0 | 0.4 | GO:0001952 | regulation of cell-matrix adhesion(GO:0001952) |
| 0.0 | 0.1 | GO:0089700 | endocardial cushion morphogenesis(GO:0003203) endocardial cushion formation(GO:0003272) protein kinase D signaling(GO:0089700) |
| 0.0 | 0.2 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.1 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.0 | 0.2 | GO:0046341 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.4 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.1 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.0 | 0.2 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 2.4 | GO:0050768 | negative regulation of neurogenesis(GO:0050768) |
| 0.0 | 0.7 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.1 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.7 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 0.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.3 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.4 | GO:0048512 | rhythmic behavior(GO:0007622) circadian behavior(GO:0048512) |
| 0.0 | 1.0 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.1 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.1 | GO:0019184 | nonribosomal peptide biosynthetic process(GO:0019184) |
| 0.0 | 0.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.4 | GO:0042159 | lipoprotein catabolic process(GO:0042159) |
| 0.0 | 0.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.3 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.1 | GO:2001240 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.2 | GO:0006543 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.0 | 0.2 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.0 | 0.6 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.0 | 0.2 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.0 | 0.4 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.9 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.2 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.1 | GO:0033628 | regulation of cell adhesion mediated by integrin(GO:0033628) |
| 0.0 | 0.3 | GO:0002761 | regulation of myeloid leukocyte differentiation(GO:0002761) |
| 0.0 | 0.3 | GO:0046113 | pyrimidine nucleobase catabolic process(GO:0006208) nucleobase catabolic process(GO:0046113) |
| 0.0 | 0.1 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.3 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.3 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.2 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.0 | 0.6 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.4 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.1 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.0 | 0.2 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.5 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.0 | 0.4 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.0 | GO:0000472 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.0 | 0.3 | GO:0097178 | ruffle organization(GO:0031529) ruffle assembly(GO:0097178) |
| 0.0 | 0.3 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.0 | 0.3 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.0 | 0.2 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.0 | 0.3 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.1 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.1 | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) negative regulation of ERBB signaling pathway(GO:1901185) |
| 0.0 | 0.8 | GO:0042738 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.2 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.2 | GO:0050795 | regulation of behavior(GO:0050795) |
| 0.0 | 0.7 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 0.5 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 1.1 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.4 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.0 | 0.2 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.2 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 0.1 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.8 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.1 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.2 | GO:0061726 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.0 | 0.1 | GO:0036268 | swimming(GO:0036268) |
| 0.0 | 0.3 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.2 | GO:0015858 | nucleoside transport(GO:0015858) nucleoside transmembrane transport(GO:1901642) |
| 0.0 | 0.5 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.1 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.0 | 0.1 | GO:0060541 | respiratory system development(GO:0060541) |
| 0.0 | 0.2 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.1 | GO:1902946 | protein localization to endosome(GO:0036010) positive regulation of receptor-mediated endocytosis(GO:0048260) protein localization to early endosome(GO:1902946) |
| 0.0 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.3 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.0 | 0.1 | GO:0046294 | formaldehyde metabolic process(GO:0046292) formaldehyde catabolic process(GO:0046294) |
| 0.0 | 0.1 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.1 | 0.8 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 1.3 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 0.4 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.1 | 0.7 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 0.3 | GO:0030062 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.1 | 0.5 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.5 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 3.8 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 0.2 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.1 | 0.7 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 0.2 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 2.4 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.7 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.5 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.7 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.3 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.2 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.5 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.3 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.1 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 0.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.9 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.4 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.2 | GO:0071914 | prominosome(GO:0071914) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.4 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 1.4 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.4 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.7 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.3 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 1.6 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.8 | GO:0022626 | cytosolic ribosome(GO:0022626) cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.5 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.2 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.8 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.2 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.9 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.6 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.7 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.7 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 1.4 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.1 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.2 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.2 | 0.6 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.2 | 0.6 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.2 | 0.5 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.2 | 0.8 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.2 | 2.3 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 0.9 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.4 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 0.5 | GO:0004772 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.1 | 0.8 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.1 | 0.9 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.1 | 0.6 | GO:0004645 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 0.6 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.1 | 0.3 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.1 | 0.5 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 0.4 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.1 | 0.5 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.1 | 0.4 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.1 | 0.3 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.1 | 0.5 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.3 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 0.5 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.4 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 0.7 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.3 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 0.3 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.5 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 0.4 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.1 | 0.5 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.7 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.1 | 0.2 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.1 | 0.2 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.2 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.2 | GO:0031843 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.0 | 0.2 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.1 | GO:0038046 | enkephalin receptor activity(GO:0038046) |
| 0.0 | 0.4 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.6 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.2 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 1.1 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.3 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.3 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.2 | GO:0016521 | pituitary adenylate cyclase activating polypeptide activity(GO:0016521) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.2 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.3 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.4 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 1.1 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.2 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.0 | 0.1 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.0 | 0.4 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.2 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.1 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.0 | 0.2 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.3 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 5.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.7 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.4 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.4 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.1 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.3 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.3 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.3 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.2 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.0 | 0.7 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.2 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 0.0 | 0.1 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.0 | 0.1 | GO:0070735 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 0.0 | 0.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.0 | 0.1 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 0.2 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.5 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.2 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 0.1 | GO:0032574 | 5'-3' RNA helicase activity(GO:0032574) |
| 0.0 | 0.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0072545 | tyrosine binding(GO:0072545) |
| 0.0 | 0.1 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.3 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 0.1 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.7 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.2 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.9 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.2 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.0 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.0 | 0.1 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.0 | 0.3 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.4 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 1.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.3 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.3 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.1 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.0 | 0.7 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.1 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.0 | 1.2 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.2 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.2 | GO:0019870 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.6 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.0 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.8 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.1 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.1 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.1 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.0 | 0.1 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.2 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.2 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.2 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.5 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.2 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.5 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.4 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 0.5 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.5 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.1 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.5 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.4 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.4 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 1.0 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 0.5 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 0.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 0.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 0.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.4 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.4 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.2 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.8 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 4.6 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.5 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.1 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.2 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 1.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.6 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.8 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 0.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.6 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.5 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.2 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.2 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.2 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.3 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.1 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.9 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.4 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |