Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
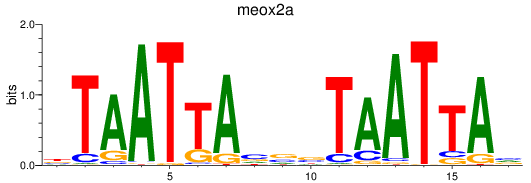
Results for meox2a
Z-value: 0.73
Transcription factors associated with meox2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
meox2a
|
ENSDARG00000040911 | mesenchyme homeobox 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| meox2a | dr11_v1_chr15_-_34458495_34458495 | 0.65 | 5.6e-02 | Click! |
Activity profile of meox2a motif
Sorted Z-values of meox2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_9669717 | 1.36 |
ENSDART00000004604
|
si:dkey-153k10.9
|
si:dkey-153k10.9 |
| chr6_+_55032439 | 1.07 |
ENSDART00000164232
ENSDART00000158845 ENSDART00000157584 ENSDART00000026359 ENSDART00000122794 ENSDART00000183742 |
mybphb
|
myosin binding protein Hb |
| chr22_-_10459880 | 1.04 |
ENSDART00000064801
|
ogn
|
osteoglycin |
| chr21_+_26390549 | 1.00 |
ENSDART00000185643
|
tmsb
|
thymosin, beta |
| chr19_-_42607451 | 0.96 |
ENSDART00000004392
|
fkbp9
|
FK506 binding protein 9 |
| chr19_+_12915498 | 0.95 |
ENSDART00000132892
|
cthrc1a
|
collagen triple helix repeat containing 1a |
| chr12_-_26407092 | 0.95 |
ENSDART00000178687
|
myoz1b
|
myozenin 1b |
| chr25_+_20089986 | 0.88 |
ENSDART00000143441
ENSDART00000184073 |
tnni4b.2
|
troponin I4b, tandem duplicate 2 |
| chr23_-_23401305 | 0.76 |
ENSDART00000078936
|
her9
|
hairy-related 9 |
| chr1_-_12278056 | 0.74 |
ENSDART00000139336
ENSDART00000137463 |
cplx2l
|
complexin 2, like |
| chr1_+_17593392 | 0.74 |
ENSDART00000078889
|
helt
|
helt bHLH transcription factor |
| chr6_-_58764672 | 0.73 |
ENSDART00000154322
|
soat2
|
sterol O-acyltransferase 2 |
| chr19_-_32150078 | 0.73 |
ENSDART00000134934
ENSDART00000186410 ENSDART00000181780 |
pag1
|
phosphoprotein membrane anchor with glycosphingolipid microdomains 1 |
| chr25_+_20091021 | 0.72 |
ENSDART00000187545
ENSDART00000053265 |
tnni4b.2
|
troponin I4b, tandem duplicate 2 |
| chr9_-_22310919 | 0.68 |
ENSDART00000108719
|
crygm2d10
|
crystallin, gamma M2d10 |
| chr9_-_22245572 | 0.68 |
ENSDART00000114943
|
crygm2d4
|
crystallin, gamma M2d4 |
| chr17_+_17955063 | 0.65 |
ENSDART00000104999
|
ccdc85ca
|
coiled-coil domain containing 85C, a |
| chr12_+_13118540 | 0.65 |
ENSDART00000077840
ENSDART00000127870 |
cmn
|
calymmin |
| chr3_+_28953274 | 0.63 |
ENSDART00000133528
ENSDART00000103602 |
lgals2a
|
lectin, galactoside-binding, soluble, 2a |
| chr21_-_43015383 | 0.63 |
ENSDART00000065097
|
dpysl3
|
dihydropyrimidinase-like 3 |
| chr21_+_28958471 | 0.61 |
ENSDART00000144331
ENSDART00000005929 |
ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr5_+_32222303 | 0.59 |
ENSDART00000051362
|
myhc4
|
myosin heavy chain 4 |
| chr13_+_43247936 | 0.59 |
ENSDART00000126850
ENSDART00000165331 |
smoc2
|
SPARC related modular calcium binding 2 |
| chr3_+_36424055 | 0.58 |
ENSDART00000170318
|
si:ch1073-443f11.2
|
si:ch1073-443f11.2 |
| chr5_-_67471375 | 0.56 |
ENSDART00000147009
|
si:dkey-251i10.2
|
si:dkey-251i10.2 |
| chr11_+_14622379 | 0.55 |
ENSDART00000112589
|
efna2b
|
ephrin-A2b |
| chr4_-_11737939 | 0.55 |
ENSDART00000150299
|
podxl
|
podocalyxin-like |
| chr11_-_1509773 | 0.55 |
ENSDART00000050762
|
phactr3b
|
phosphatase and actin regulator 3b |
| chr9_-_48371379 | 0.54 |
ENSDART00000115121
|
col28a2a
|
collagen, type XXVIII, alpha 2a |
| chr16_-_43344859 | 0.54 |
ENSDART00000058680
|
psma2
|
proteasome subunit alpha 2 |
| chr13_-_1408775 | 0.54 |
ENSDART00000049684
|
bag2
|
BCL2 associated athanogene 2 |
| chr3_-_41795917 | 0.53 |
ENSDART00000182662
|
grifin
|
galectin-related inter-fiber protein |
| chr16_+_43344475 | 0.52 |
ENSDART00000085282
|
mrpl32
|
mitochondrial ribosomal protein L32 |
| chr13_+_29778610 | 0.52 |
ENSDART00000132004
|
pax2a
|
paired box 2a |
| chr21_-_26071773 | 0.52 |
ENSDART00000141382
|
rab34b
|
RAB34, member RAS oncogene family b |
| chr8_+_29986265 | 0.51 |
ENSDART00000148258
|
ptch1
|
patched 1 |
| chr11_+_8565323 | 0.49 |
ENSDART00000081607
ENSDART00000141612 |
tbl1xr1a
|
transducin (beta)-like 1 X-linked receptor 1a |
| chr4_-_18595525 | 0.49 |
ENSDART00000049061
|
cdkn1ba
|
cyclin-dependent kinase inhibitor 1Ba |
| chr5_-_63509581 | 0.48 |
ENSDART00000097325
|
c5
|
complement component 5 |
| chr23_+_42482137 | 0.47 |
ENSDART00000160199
|
cyp2aa3
|
cytochrome P450, family 2, subfamily AA, polypeptide 3 |
| chr21_-_5958387 | 0.47 |
ENSDART00000130521
|
CACFD1
|
si:ch73-209e20.5 |
| chr19_+_43396861 | 0.47 |
ENSDART00000151433
|
yrk
|
Yes-related kinase |
| chr19_+_5072918 | 0.47 |
ENSDART00000037126
|
eno2
|
enolase 2 |
| chr4_-_22311610 | 0.46 |
ENSDART00000137814
|
hcls1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr13_+_19322686 | 0.46 |
ENSDART00000058036
|
emx2
|
empty spiracles homeobox 2 |
| chr12_+_30586599 | 0.46 |
ENSDART00000124920
ENSDART00000126984 |
nrap
|
nebulin-related anchoring protein |
| chr11_+_8565828 | 0.45 |
ENSDART00000126523
|
tbl1xr1a
|
transducin (beta)-like 1 X-linked receptor 1a |
| chr11_-_25418856 | 0.45 |
ENSDART00000013714
|
gata1a
|
GATA binding protein 1a |
| chr23_+_36106790 | 0.44 |
ENSDART00000128533
|
hoxc3a
|
homeobox C3a |
| chr14_+_25465346 | 0.44 |
ENSDART00000173436
|
si:dkey-280e21.3
|
si:dkey-280e21.3 |
| chr5_+_29820266 | 0.44 |
ENSDART00000146331
ENSDART00000098315 |
f11r.2
|
F11 receptor, tandem duplicate 2 |
| chr6_-_11768198 | 0.44 |
ENSDART00000183463
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr6_-_18976168 | 0.44 |
ENSDART00000170039
|
sept9b
|
septin 9b |
| chr23_-_15284757 | 0.43 |
ENSDART00000139135
|
sulf2b
|
sulfatase 2b |
| chr7_+_67467702 | 0.43 |
ENSDART00000168460
ENSDART00000170322 |
rpl13
|
ribosomal protein L13 |
| chr8_+_52619365 | 0.42 |
ENSDART00000162953
|
cyr61l2
|
cysteine-rich, angiogenic inducer, 61 like 2 |
| chr23_-_11870962 | 0.42 |
ENSDART00000143481
|
si:dkey-178k16.1
|
si:dkey-178k16.1 |
| chr2_+_55982300 | 0.41 |
ENSDART00000183903
|
nmrk2
|
nicotinamide riboside kinase 2 |
| chr6_-_58757131 | 0.40 |
ENSDART00000083582
|
soat2
|
sterol O-acyltransferase 2 |
| chr9_-_42418470 | 0.40 |
ENSDART00000144353
|
calcrla
|
calcitonin receptor-like a |
| chr5_+_29159777 | 0.40 |
ENSDART00000174702
ENSDART00000037891 |
dpp7
|
dipeptidyl-peptidase 7 |
| chr5_+_48683447 | 0.40 |
ENSDART00000008043
|
adgrv1
|
adhesion G protein-coupled receptor V1 |
| chr23_+_45538932 | 0.40 |
ENSDART00000135602
|
si:ch73-290k24.6
|
si:ch73-290k24.6 |
| chr22_+_9872323 | 0.40 |
ENSDART00000129240
|
si:dkey-253d23.4
|
si:dkey-253d23.4 |
| chr25_+_5044780 | 0.39 |
ENSDART00000153980
|
parvb
|
parvin, beta |
| chr2_+_24188502 | 0.39 |
ENSDART00000181955
ENSDART00000191706 ENSDART00000125909 |
map4l
|
microtubule associated protein 4 like |
| chr4_+_25223050 | 0.38 |
ENSDART00000138121
|
itih5
|
inter-alpha-trypsin inhibitor heavy chain family, member 5 |
| chr16_+_12836143 | 0.38 |
ENSDART00000067741
|
cacng6b
|
calcium channel, voltage-dependent, gamma subunit 6b |
| chr16_-_31754102 | 0.38 |
ENSDART00000185043
|
ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr20_+_4060839 | 0.36 |
ENSDART00000178565
|
TRIM67
|
tripartite motif containing 67 |
| chr2_+_50862527 | 0.36 |
ENSDART00000169800
ENSDART00000158847 ENSDART00000160900 |
adcyap1r1a
|
adenylate cyclase activating polypeptide 1a (pituitary) receptor type I |
| chr24_+_24831727 | 0.36 |
ENSDART00000080969
|
trim55b
|
tripartite motif containing 55b |
| chr21_+_17880511 | 0.35 |
ENSDART00000080481
|
rxraa
|
retinoid X receptor, alpha a |
| chr20_+_26966725 | 0.35 |
ENSDART00000029781
|
ahsa1a
|
AHA1, activator of heat shock protein ATPase homolog 1a |
| chr5_-_9073433 | 0.35 |
ENSDART00000099891
|
atp5meb
|
ATP synthase membrane subunit eb |
| chr14_+_11458044 | 0.34 |
ENSDART00000186425
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr12_+_18681477 | 0.34 |
ENSDART00000127981
ENSDART00000143979 |
rgs9b
|
regulator of G protein signaling 9b |
| chr14_+_22113331 | 0.34 |
ENSDART00000109759
|
tmx2a
|
thioredoxin-related transmembrane protein 2a |
| chr2_+_33382648 | 0.34 |
ENSDART00000137207
ENSDART00000098831 |
slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr16_+_23960744 | 0.33 |
ENSDART00000058965
|
apoeb
|
apolipoprotein Eb |
| chr11_-_2131280 | 0.33 |
ENSDART00000008409
|
calcoco1b
|
calcium binding and coiled-coil domain 1b |
| chr21_+_26071874 | 0.33 |
ENSDART00000003001
ENSDART00000146573 |
rpl23a
|
ribosomal protein L23a |
| chr3_-_12930217 | 0.33 |
ENSDART00000166322
|
pdgfab
|
platelet-derived growth factor alpha polypeptide b |
| chr24_-_37287792 | 0.33 |
ENSDART00000166086
|
CABZ01055522.1
|
|
| chr15_+_36966369 | 0.33 |
ENSDART00000163622
|
kirrel3l
|
kirre like nephrin family adhesion molecule 3, like |
| chr14_-_11456724 | 0.33 |
ENSDART00000110424
|
si:ch211-153b23.4
|
si:ch211-153b23.4 |
| chr1_-_58963395 | 0.32 |
ENSDART00000130415
|
CABZ01115881.1
|
|
| chr19_-_38611814 | 0.32 |
ENSDART00000151958
|
col16a1
|
collagen, type XVI, alpha 1 |
| chr23_-_45539306 | 0.32 |
ENSDART00000161913
|
CU681841.1
|
|
| chr1_-_10647307 | 0.32 |
ENSDART00000103548
|
si:dkey-31e10.1
|
si:dkey-31e10.1 |
| chr15_-_2188332 | 0.32 |
ENSDART00000138941
ENSDART00000009564 |
shox2
|
short stature homeobox 2 |
| chr3_+_36284986 | 0.32 |
ENSDART00000059533
|
wipi1
|
WD repeat domain, phosphoinositide interacting 1 |
| chr5_+_51026563 | 0.31 |
ENSDART00000050988
|
gcnt4a
|
glucosaminyl (N-acetyl) transferase 4, core 2, a |
| chr24_+_23742690 | 0.31 |
ENSDART00000130162
|
TCF24
|
transcription factor 24 |
| chr16_+_5184402 | 0.31 |
ENSDART00000156685
|
soga3a
|
SOGA family member 3a |
| chr17_-_21441464 | 0.31 |
ENSDART00000031490
|
vax1
|
ventral anterior homeobox 1 |
| chr20_-_8094718 | 0.31 |
ENSDART00000122902
|
si:ch211-232i5.3
|
si:ch211-232i5.3 |
| chr12_-_36740781 | 0.30 |
ENSDART00000105484
|
si:ch211-216b21.2
|
si:ch211-216b21.2 |
| chr4_-_8014463 | 0.30 |
ENSDART00000036153
|
ccdc3a
|
coiled-coil domain containing 3a |
| chr12_-_28570989 | 0.30 |
ENSDART00000008010
|
pdk2a
|
pyruvate dehydrogenase kinase, isozyme 2a |
| chr16_-_27174373 | 0.30 |
ENSDART00000166681
|
frrs1l
|
ferric-chelate reductase 1-like |
| chr1_+_25801648 | 0.30 |
ENSDART00000129471
|
gucy1b1
|
guanylate cyclase 1 soluble subunit beta 1 |
| chr5_+_65871688 | 0.30 |
ENSDART00000175217
|
FAM163B (1 of many)
|
family with sequence similarity 163 member B |
| chr5_+_29160132 | 0.29 |
ENSDART00000088827
|
dpp7
|
dipeptidyl-peptidase 7 |
| chr9_-_1703761 | 0.29 |
ENSDART00000144822
ENSDART00000137210 ENSDART00000135273 |
hnrnpa3
|
heterogeneous nuclear ribonucleoprotein A3 |
| chr21_-_26071142 | 0.29 |
ENSDART00000004740
|
rab34b
|
RAB34, member RAS oncogene family b |
| chr12_-_48477031 | 0.29 |
ENSDART00000105176
|
ndufb8
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 8 |
| chr21_-_5205617 | 0.29 |
ENSDART00000145554
ENSDART00000045284 |
rpl37
|
ribosomal protein L37 |
| chr16_+_34479064 | 0.29 |
ENSDART00000159965
|
si:ch211-255i3.4
|
si:ch211-255i3.4 |
| chr25_-_18140305 | 0.28 |
ENSDART00000180222
|
kitlga
|
kit ligand a |
| chr12_+_17504559 | 0.28 |
ENSDART00000020628
|
cyth3a
|
cytohesin 3a |
| chr21_-_5881344 | 0.28 |
ENSDART00000009241
|
rpl35
|
ribosomal protein L35 |
| chr6_+_32497493 | 0.28 |
ENSDART00000184819
|
kank4
|
KN motif and ankyrin repeat domains 4 |
| chr25_+_3677650 | 0.28 |
ENSDART00000154348
|
prnprs3
|
prion protein, related sequence 3 |
| chr1_-_10647484 | 0.27 |
ENSDART00000164541
ENSDART00000188958 ENSDART00000190904 |
si:dkey-31e10.1
|
si:dkey-31e10.1 |
| chr4_+_391297 | 0.27 |
ENSDART00000030215
|
rpl18a
|
ribosomal protein L18a |
| chr11_-_30158191 | 0.27 |
ENSDART00000155278
ENSDART00000156121 |
scml2
|
Scm polycomb group protein like 2 |
| chr10_-_13239367 | 0.26 |
ENSDART00000001253
|
si:busm1-57f23.1
|
si:busm1-57f23.1 |
| chr10_+_21576909 | 0.26 |
ENSDART00000168604
ENSDART00000166533 |
pcdh1a3
|
protocadherin 1 alpha 3 |
| chr5_+_29160324 | 0.26 |
ENSDART00000137324
|
dpp7
|
dipeptidyl-peptidase 7 |
| chr17_-_49978986 | 0.26 |
ENSDART00000154728
|
col12a1a
|
collagen, type XII, alpha 1a |
| chr18_-_17030870 | 0.26 |
ENSDART00000079817
|
has3
|
hyaluronan synthase 3 |
| chr25_-_16755340 | 0.26 |
ENSDART00000124729
ENSDART00000110859 |
ribc2
|
RIB43A domain with coiled-coils 2 |
| chr25_-_8030425 | 0.25 |
ENSDART00000014964
|
camk1db
|
calcium/calmodulin-dependent protein kinase 1Db |
| chr8_-_13972626 | 0.25 |
ENSDART00000144296
|
serping1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr1_+_21725821 | 0.25 |
ENSDART00000132602
|
pax5
|
paired box 5 |
| chr10_-_20524592 | 0.25 |
ENSDART00000185048
|
ddhd2
|
DDHD domain containing 2 |
| chr21_-_41025340 | 0.25 |
ENSDART00000148231
|
plac8l1
|
PLAC8-like 1 |
| chr20_+_1960092 | 0.25 |
ENSDART00000191892
|
CABZ01103860.1
|
|
| chr13_+_30903816 | 0.25 |
ENSDART00000191727
|
ercc6
|
excision repair cross-complementation group 6 |
| chr20_-_4766645 | 0.25 |
ENSDART00000147071
ENSDART00000152398 |
ak7a
|
adenylate kinase 7a |
| chr16_+_16969060 | 0.25 |
ENSDART00000182819
ENSDART00000191876 |
si:ch211-120k19.1
rpl18
|
si:ch211-120k19.1 ribosomal protein L18 |
| chr13_-_37636147 | 0.25 |
ENSDART00000144065
|
si:dkey-188i13.8
|
si:dkey-188i13.8 |
| chr16_+_54780544 | 0.25 |
ENSDART00000126646
|
si:zfos-1192g2.3
|
si:zfos-1192g2.3 |
| chr7_-_9803154 | 0.25 |
ENSDART00000055593
|
aldh1a3
|
aldehyde dehydrogenase 1 family, member A3 |
| chr1_+_55293424 | 0.25 |
ENSDART00000152464
|
zgc:172106
|
zgc:172106 |
| chr15_+_14202114 | 0.24 |
ENSDART00000169369
|
si:dkey-243k1.3
|
si:dkey-243k1.3 |
| chr6_-_10753504 | 0.24 |
ENSDART00000045299
ENSDART00000165645 |
ola1
|
Obg-like ATPase 1 |
| chr10_-_35236949 | 0.24 |
ENSDART00000145804
|
ypel2a
|
yippee-like 2a |
| chr10_-_20339113 | 0.24 |
ENSDART00000144378
ENSDART00000098600 |
zmat4b
|
zinc finger, matrin-type 4b |
| chr14_-_2001386 | 0.24 |
ENSDART00000170346
ENSDART00000163975 |
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr22_-_12746539 | 0.24 |
ENSDART00000175374
|
plcd4a
|
phospholipase C, delta 4a |
| chr14_+_36869273 | 0.24 |
ENSDART00000111674
|
F2RL3
|
si:ch211-132p1.2 |
| chr15_+_28355023 | 0.24 |
ENSDART00000122159
|
si:dkey-118k5.3
|
si:dkey-118k5.3 |
| chr7_+_25858380 | 0.24 |
ENSDART00000148780
ENSDART00000079218 |
mtmr1a
|
myotubularin related protein 1a |
| chr1_+_51721851 | 0.24 |
ENSDART00000040397
|
prdx2
|
peroxiredoxin 2 |
| chrM_+_11009 | 0.23 |
ENSDART00000093617
|
mt-nd4l
|
NADH dehydrogenase 4L, mitochondrial |
| chr1_+_45085194 | 0.23 |
ENSDART00000193863
|
si:ch211-151p13.8
|
si:ch211-151p13.8 |
| chr15_+_9053059 | 0.23 |
ENSDART00000012039
|
ppm1na
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Na (putative) |
| chr15_-_2632891 | 0.23 |
ENSDART00000081840
|
cldnj
|
claudin j |
| chr11_+_43431289 | 0.23 |
ENSDART00000192526
|
slc29a1b
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1b |
| chr10_+_8730988 | 0.23 |
ENSDART00000145596
|
itga1
|
integrin, alpha 1 |
| chr23_-_45705525 | 0.23 |
ENSDART00000148959
|
ednrab
|
endothelin receptor type Ab |
| chr1_-_44701313 | 0.22 |
ENSDART00000193926
|
si:dkey-28b4.8
|
si:dkey-28b4.8 |
| chr12_+_28854410 | 0.22 |
ENSDART00000152991
|
nfe2l1b
|
nuclear factor, erythroid 2-like 1b |
| chr2_-_51644044 | 0.22 |
ENSDART00000157899
|
dad1
|
defender against cell death 1 |
| chr19_+_1831911 | 0.22 |
ENSDART00000166653
|
ptk2aa
|
protein tyrosine kinase 2aa |
| chr22_+_17205608 | 0.22 |
ENSDART00000181267
|
rab3b
|
RAB3B, member RAS oncogene family |
| chr16_+_11834516 | 0.22 |
ENSDART00000146611
|
cxcr3.3
|
chemokine (C-X-C motif) receptor 3, tandem duplicate 3 |
| chr19_-_10810006 | 0.22 |
ENSDART00000151157
|
si:dkey-3n22.9
|
si:dkey-3n22.9 |
| chr15_+_33865312 | 0.21 |
ENSDART00000166141
|
tekt1
|
tektin 1 |
| chr25_-_10565006 | 0.21 |
ENSDART00000130608
ENSDART00000190212 |
galn
|
galanin/GMAP prepropeptide |
| chr20_-_22476255 | 0.21 |
ENSDART00000103510
|
pdgfra
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr7_+_48705227 | 0.21 |
ENSDART00000174034
|
AL929208.1
|
|
| chr6_-_33023745 | 0.21 |
ENSDART00000156211
|
adcyap1r1b
|
adenylate cyclase activating polypeptide 1b (pituitary) receptor type I |
| chr14_-_30387894 | 0.21 |
ENSDART00000176136
|
slc7a2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
| chr9_-_31596016 | 0.21 |
ENSDART00000142289
|
nalcn
|
sodium leak channel, non-selective |
| chr24_-_40860603 | 0.21 |
ENSDART00000188032
|
CU633479.7
|
|
| chr10_+_4874542 | 0.21 |
ENSDART00000101431
|
palm2
|
paralemmin 2 |
| chr12_+_13652361 | 0.21 |
ENSDART00000182757
ENSDART00000152689 |
oplah
|
5-oxoprolinase, ATP-hydrolysing |
| chr8_-_10932206 | 0.21 |
ENSDART00000124313
|
nr1h5
|
nuclear receptor subfamily 1, group H, member 5 |
| chr8_+_34624025 | 0.21 |
ENSDART00000180772
|
BX247865.2
|
|
| chr3_+_59784632 | 0.21 |
ENSDART00000084729
|
pecam1
|
platelet/endothelial cell adhesion molecule 1 |
| chr7_+_8456999 | 0.21 |
ENSDART00000172880
|
jac4
|
jacalin 4 |
| chr16_+_14588141 | 0.21 |
ENSDART00000140469
ENSDART00000059984 ENSDART00000167411 ENSDART00000133566 |
deptor
|
DEP domain containing MTOR-interacting protein |
| chr25_+_31267268 | 0.21 |
ENSDART00000181239
|
tnni2a.3
|
troponin I type 2a (skeletal, fast), tandem duplicate 3 |
| chr9_-_8661436 | 0.21 |
ENSDART00000130442
|
col4a1
|
collagen, type IV, alpha 1 |
| chr10_+_16092671 | 0.21 |
ENSDART00000182761
ENSDART00000154835 |
megf10
|
multiple EGF-like-domains 10 |
| chr2_-_30324610 | 0.20 |
ENSDART00000185422
|
jph1b
|
junctophilin 1b |
| chr25_+_13731542 | 0.20 |
ENSDART00000161012
|
ccdc135
|
coiled-coil domain containing 135 |
| chr2_+_10709557 | 0.20 |
ENSDART00000183118
ENSDART00000109723 |
evi5a
|
ecotropic viral integration site 5a |
| chr1_+_56463494 | 0.20 |
ENSDART00000097964
|
zgc:171452
|
zgc:171452 |
| chr25_+_26895394 | 0.20 |
ENSDART00000155820
|
si:dkey-42p14.3
|
si:dkey-42p14.3 |
| chr2_+_35854242 | 0.20 |
ENSDART00000134918
|
dhx9
|
DEAH (Asp-Glu-Ala-His) box helicase 9 |
| chr3_-_39152478 | 0.20 |
ENSDART00000154550
|
si:dkeyp-57f11.2
|
si:dkeyp-57f11.2 |
| chr14_-_33218418 | 0.20 |
ENSDART00000163046
|
si:dkey-31j3.11
|
si:dkey-31j3.11 |
| chr6_+_27062512 | 0.19 |
ENSDART00000168277
ENSDART00000149429 |
boka
|
BOK, BCL2 family apoptosis regulator a |
| chr16_-_51888952 | 0.19 |
ENSDART00000186407
ENSDART00000175435 |
rpl28
|
ribosomal protein L28 |
| chr11_+_2506516 | 0.19 |
ENSDART00000130886
ENSDART00000189767 |
NABP2
|
si:ch73-190f16.2 |
| chr18_+_7286788 | 0.19 |
ENSDART00000022998
|
ANO2 (1 of many)
|
si:ch73-86n2.1 |
| chr13_+_4368504 | 0.19 |
ENSDART00000108935
|
sft2d1
|
SFT2 domain containing 1 |
| chr22_-_1245226 | 0.19 |
ENSDART00000161292
|
ampd2a
|
adenosine monophosphate deaminase 2a |
| chr7_+_69905307 | 0.18 |
ENSDART00000035907
|
sec24d
|
SEC24 homolog D, COPII coat complex component |
| chr19_-_2861444 | 0.18 |
ENSDART00000169053
|
clec3bb
|
C-type lectin domain family 3, member Bb |
| chr7_-_71758307 | 0.18 |
ENSDART00000161067
ENSDART00000165253 |
myom1b
|
myomesin 1b |
| chr3_-_58116314 | 0.18 |
ENSDART00000154901
|
si:ch211-256e16.6
|
si:ch211-256e16.6 |
| chr10_+_31248036 | 0.18 |
ENSDART00000193574
|
robo4
|
roundabout, axon guidance receptor, homolog 4 (Drosophila) |
| chr5_+_24245682 | 0.17 |
ENSDART00000049003
|
atp6v1aa
|
ATPase H+ transporting V1 subunit Aa |
| chr12_-_3753131 | 0.17 |
ENSDART00000129668
|
fam57bb
|
family with sequence similarity 57, member Bb |
| chr16_+_20161805 | 0.17 |
ENSDART00000192146
|
c16h2orf66
|
chromosome 16 C2orf66 homolog |
Network of associatons between targets according to the STRING database.
First level regulatory network of meox2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.2 | 0.5 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.1 | 1.2 | GO:0060118 | vestibular receptor cell differentiation(GO:0060114) vestibular receptor cell development(GO:0060118) |
| 0.1 | 1.2 | GO:0034433 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.1 | 0.4 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 0.4 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.1 | 0.3 | GO:0071831 | high-density lipoprotein particle assembly(GO:0034380) chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 0.1 | 0.8 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.1 | 0.3 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.1 | 0.3 | GO:2000257 | regulation of protein activation cascade(GO:2000257) |
| 0.1 | 0.3 | GO:0060092 | regulation of synaptic transmission, glycinergic(GO:0060092) |
| 0.1 | 0.2 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) |
| 0.1 | 0.2 | GO:1904088 | regulation of convergent extension involved in axis elongation(GO:1901232) positive regulation of epiboly involved in gastrulation with mouth forming second(GO:1904088) |
| 0.1 | 0.3 | GO:0090259 | regulation of retinal ganglion cell axon guidance(GO:0090259) |
| 0.1 | 0.4 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.1 | 0.5 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.1 | 0.3 | GO:0099543 | trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.1 | 0.5 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.1 | 0.1 | GO:0033630 | positive regulation of cell adhesion mediated by integrin(GO:0033630) |
| 0.1 | 0.2 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.1 | 0.2 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.1 | 0.5 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.1 | 0.5 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.1 | 0.3 | GO:0021731 | trigeminal motor nucleus development(GO:0021731) |
| 0.1 | 0.6 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.3 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
| 0.0 | 0.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.3 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.2 | GO:0031174 | lifelong otolith mineralization(GO:0031174) |
| 0.0 | 0.2 | GO:0003319 | cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) |
| 0.0 | 0.6 | GO:0090303 | positive regulation of wound healing(GO:0090303) |
| 0.0 | 0.2 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) heart rudiment formation(GO:0003315) |
| 0.0 | 2.1 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.6 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.2 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.0 | 0.2 | GO:0015862 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.1 | GO:0099538 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 0.3 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.0 | 0.2 | GO:0090467 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.3 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.3 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.4 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.0 | 0.4 | GO:0035860 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.2 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.0 | 0.2 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.0 | 0.6 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.0 | 0.4 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.2 | GO:0000303 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.1 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.0 | 0.1 | GO:0046084 | adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.6 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.3 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.3 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 0.7 | GO:0050868 | negative regulation of T cell activation(GO:0050868) negative regulation of leukocyte cell-cell adhesion(GO:1903038) |
| 0.0 | 0.4 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.5 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.0 | 0.6 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.0 | 0.1 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.9 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.1 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 | 0.6 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.3 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.2 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.2 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0015990 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) electron transport coupled proton transport(GO:0015990) |
| 0.0 | 0.3 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.1 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.0 | 0.1 | GO:0039528 | cytoplasmic pattern recognition receptor signaling pathway in response to virus(GO:0039528) cellular response to virus(GO:0098586) |
| 0.0 | 0.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.4 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.9 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.5 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 0.3 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.3 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.1 | GO:0071423 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.0 | 0.1 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.2 | GO:0030317 | sperm motility(GO:0030317) |
| 0.0 | 0.3 | GO:0070570 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.0 | 0.1 | GO:1903672 | positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.0 | 0.2 | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.0 | 0.1 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.6 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.1 | GO:0031643 | positive regulation of myelination(GO:0031643) positive regulation of neurological system process(GO:0031646) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.4 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.1 | 0.3 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.1 | 0.5 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.5 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 0.4 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 2.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.5 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.3 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.7 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 1.8 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.2 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 2.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.8 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.7 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.5 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.9 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 0.0 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 0.4 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.2 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.5 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.4 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.2 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0004772 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.2 | 0.5 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.1 | 0.4 | GO:0001635 | adrenomedullin receptor activity(GO:0001605) calcitonin gene-related peptide receptor activity(GO:0001635) |
| 0.1 | 0.5 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.1 | 0.3 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 1.1 | GO:0051373 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.1 | 0.6 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.4 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.5 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 0.6 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.5 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 0.4 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.1 | 0.5 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.4 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.1 | 0.5 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 1.0 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 0.2 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.6 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 0.2 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.0 | 0.2 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 1.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.3 | GO:0030291 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) protein serine/threonine kinase inhibitor activity(GO:0030291) |
| 0.0 | 0.4 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.3 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.0 | 0.3 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.3 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.1 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.2 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.2 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 0.4 | GO:0010851 | cyclase regulator activity(GO:0010851) |
| 0.0 | 0.2 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.2 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.1 | GO:0035671 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) enone reductase activity(GO:0035671) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 0.1 | GO:0031530 | gonadotropin hormone-releasing hormone activity(GO:0005183) gonadotropin-releasing hormone receptor binding(GO:0031530) |
| 0.0 | 0.3 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.6 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0098639 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.3 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.2 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.2 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.3 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.1 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.0 | 0.2 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.2 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.2 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.5 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.3 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.0 | 0.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.0 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.1 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.1 | GO:0015131 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.4 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.2 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.2 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 2.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.9 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.0 | 0.1 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 0.8 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.6 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 1.1 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.1 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.2 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.4 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.2 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.1 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.3 | NABA COLLAGENS | Genes encoding collagen proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.4 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.1 | 0.7 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 1.0 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 0.6 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.3 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.2 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 1.0 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 2.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.5 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.1 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.6 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.1 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.2 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.0 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.1 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.2 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |