Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
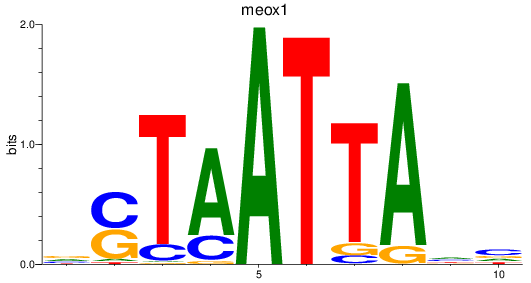
Results for meox1
Z-value: 0.99
Transcription factors associated with meox1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
meox1
|
ENSDARG00000007891 | mesenchyme homeobox 1 |
|
meox1
|
ENSDARG00000115382 | mesenchyme homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| meox1 | dr11_v1_chr12_+_27462225_27462225 | -0.95 | 7.8e-05 | Click! |
Activity profile of meox1 motif
Sorted Z-values of meox1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_-_40708537 | 3.04 |
ENSDART00000077577
|
si:ch211-132b12.8
|
si:ch211-132b12.8 |
| chr2_-_15324837 | 2.84 |
ENSDART00000015655
|
tecrl2b
|
trans-2,3-enoyl-CoA reductase-like 2b |
| chr11_-_44801968 | 2.63 |
ENSDART00000161846
|
map1lc3c
|
microtubule-associated protein 1 light chain 3 gamma |
| chr20_-_6532462 | 2.55 |
ENSDART00000054653
|
mcm3l
|
MCM3 minichromosome maintenance deficient 3 (S. cerevisiae), like |
| chr2_-_50225411 | 2.38 |
ENSDART00000147117
ENSDART00000000042 |
mcm6l
|
MCM6 minichromosome maintenance deficient 6, like |
| chr8_-_23780334 | 2.09 |
ENSDART00000145179
ENSDART00000145894 |
zgc:195245
|
zgc:195245 |
| chr10_-_34002185 | 2.01 |
ENSDART00000046599
|
zar1l
|
zygote arrest 1-like |
| chr24_+_1023839 | 1.99 |
ENSDART00000082526
|
zgc:111976
|
zgc:111976 |
| chr1_-_23308225 | 1.99 |
ENSDART00000137567
ENSDART00000008201 |
smim14
|
small integral membrane protein 14 |
| chr11_-_1550709 | 1.98 |
ENSDART00000110097
|
si:ch73-303b9.1
|
si:ch73-303b9.1 |
| chr9_-_35633827 | 1.98 |
ENSDART00000077745
|
zp2l1
|
zona pellucida glycoprotein 2, like 1 |
| chr6_+_28208973 | 1.84 |
ENSDART00000171216
ENSDART00000171377 ENSDART00000167389 ENSDART00000166988 |
LSM2 (1 of many)
|
si:ch73-14h10.2 |
| chr10_-_21545091 | 1.80 |
ENSDART00000029122
ENSDART00000132207 |
zgc:165539
|
zgc:165539 |
| chr23_+_2728095 | 1.77 |
ENSDART00000066086
|
zgc:114123
|
zgc:114123 |
| chr10_-_21362071 | 1.73 |
ENSDART00000125167
|
avd
|
avidin |
| chr10_-_21362320 | 1.67 |
ENSDART00000189789
|
avd
|
avidin |
| chr4_-_77140030 | 1.67 |
ENSDART00000174332
ENSDART00000190394 ENSDART00000189208 |
CU467646.5
CU467646.7
|
|
| chr4_-_77125693 | 1.57 |
ENSDART00000174256
|
CU467646.3
|
|
| chr16_-_42056137 | 1.52 |
ENSDART00000102798
|
zp3d.2
|
zona pellucida glycoprotein 3d tandem duplicate 2 |
| chr20_-_23426339 | 1.50 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr9_-_38021889 | 1.48 |
ENSDART00000183482
ENSDART00000124333 |
adcy5
|
adenylate cyclase 5 |
| chr22_+_17828267 | 1.44 |
ENSDART00000136016
|
hapln4
|
hyaluronan and proteoglycan link protein 4 |
| chr6_-_43283122 | 1.35 |
ENSDART00000186022
|
frmd4ba
|
FERM domain containing 4Ba |
| chr10_-_44560165 | 1.34 |
ENSDART00000181217
ENSDART00000076084 |
npm2b
|
nucleophosmin/nucleoplasmin, 2b |
| chr11_-_6452444 | 1.27 |
ENSDART00000137879
ENSDART00000134957 ENSDART00000004483 |
larp6b
|
La ribonucleoprotein domain family, member 6b |
| chr12_-_33357655 | 1.22 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr2_-_26596794 | 1.20 |
ENSDART00000134685
ENSDART00000056787 |
zgc:113691
|
zgc:113691 |
| chr2_+_6253246 | 1.12 |
ENSDART00000058256
ENSDART00000076700 |
zp3b
|
zona pellucida glycoprotein 3b |
| chr10_+_6884627 | 1.08 |
ENSDART00000125262
ENSDART00000121729 ENSDART00000105384 |
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr4_+_13586689 | 1.07 |
ENSDART00000067161
ENSDART00000138201 |
tnpo3
|
transportin 3 |
| chr9_+_22656976 | 1.02 |
ENSDART00000136249
ENSDART00000139270 |
si:dkey-189g17.2
|
si:dkey-189g17.2 |
| chr24_+_40905100 | 1.02 |
ENSDART00000167854
|
scn12ab
|
sodium channel, voltage gated, type XII, alpha b |
| chr1_+_51191049 | 1.01 |
ENSDART00000132244
ENSDART00000014970 ENSDART00000132141 |
btbd3a
|
BTB (POZ) domain containing 3a |
| chr17_+_49500820 | 1.01 |
ENSDART00000170306
|
AREL1
|
apoptosis resistant E3 ubiquitin protein ligase 1 |
| chr11_+_31864921 | 1.00 |
ENSDART00000180252
|
diaph3
|
diaphanous-related formin 3 |
| chr5_-_30074332 | 0.97 |
ENSDART00000147963
|
bco2a
|
beta-carotene oxygenase 2a |
| chr6_-_2134581 | 0.97 |
ENSDART00000175478
|
vstm2l
|
V-set and transmembrane domain containing 2 like |
| chr20_-_45060241 | 0.94 |
ENSDART00000185227
|
klhl29
|
kelch-like family member 29 |
| chr23_-_31913069 | 0.94 |
ENSDART00000135526
|
mtfr2
|
mitochondrial fission regulator 2 |
| chr19_-_18127808 | 0.92 |
ENSDART00000108627
|
snx10a
|
sorting nexin 10a |
| chr19_-_18127629 | 0.90 |
ENSDART00000187722
|
snx10a
|
sorting nexin 10a |
| chr14_-_33481428 | 0.90 |
ENSDART00000147059
ENSDART00000140001 ENSDART00000124242 ENSDART00000164836 ENSDART00000190104 ENSDART00000186833 ENSDART00000180873 |
lamp2
|
lysosomal-associated membrane protein 2 |
| chr22_-_21897203 | 0.89 |
ENSDART00000158501
ENSDART00000105566 ENSDART00000136795 |
gna11a
|
guanine nucleotide binding protein (G protein), alpha 11a (Gq class) |
| chr14_+_34490445 | 0.85 |
ENSDART00000132193
ENSDART00000148044 |
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr20_-_38787047 | 0.85 |
ENSDART00000152913
ENSDART00000153430 |
dnajc5ga
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma a |
| chr1_-_53468160 | 0.85 |
ENSDART00000143349
|
zgc:66455
|
zgc:66455 |
| chr20_-_38787341 | 0.81 |
ENSDART00000136771
|
dnajc5ga
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma a |
| chr13_+_38814521 | 0.81 |
ENSDART00000110976
|
col19a1
|
collagen, type XIX, alpha 1 |
| chr10_-_35257458 | 0.80 |
ENSDART00000143890
ENSDART00000139107 ENSDART00000082445 |
prr11
|
proline rich 11 |
| chr19_-_5103141 | 0.80 |
ENSDART00000150952
|
tpi1a
|
triosephosphate isomerase 1a |
| chr15_+_31344472 | 0.79 |
ENSDART00000146695
ENSDART00000159182 ENSDART00000060125 |
or107-1
|
odorant receptor, family D, subfamily 107, member 1 |
| chr12_+_46582727 | 0.79 |
ENSDART00000149326
|
ush1gb
|
Usher syndrome 1Gb (autosomal recessive) |
| chr15_-_34930727 | 0.78 |
ENSDART00000179723
|
dhx16
|
DEAH (Asp-Glu-Ala-His) box polypeptide 16 |
| chr7_+_69459759 | 0.77 |
ENSDART00000160500
|
ctdnep1b
|
CTD nuclear envelope phosphatase 1b |
| chr8_-_30338872 | 0.77 |
ENSDART00000137583
|
dock8
|
dedicator of cytokinesis 8 |
| chr6_+_4370935 | 0.76 |
ENSDART00000192368
|
rbm26
|
RNA binding motif protein 26 |
| chr19_+_8612839 | 0.76 |
ENSDART00000144925
|
snx27a
|
sorting nexin family member 27a |
| chr17_+_16046132 | 0.75 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr12_+_30367371 | 0.75 |
ENSDART00000153364
|
ccdc186
|
si:ch211-225b10.4 |
| chr25_+_35891342 | 0.75 |
ENSDART00000147093
|
lsm14aa
|
LSM14A mRNA processing body assembly factor a |
| chr6_+_21001264 | 0.74 |
ENSDART00000044519
ENSDART00000151278 |
cx44.2
|
connexin 44.2 |
| chr11_-_34219211 | 0.72 |
ENSDART00000098472
|
tmem44
|
transmembrane protein 44 |
| chr5_+_40837539 | 0.71 |
ENSDART00000188279
|
si:dkey-3h3.3
|
si:dkey-3h3.3 |
| chr4_+_13586455 | 0.71 |
ENSDART00000187230
|
tnpo3
|
transportin 3 |
| chr24_+_39518774 | 0.69 |
ENSDART00000132939
|
dcun1d3
|
defective in cullin neddylation 1 domain containing 3 |
| chr6_+_54576520 | 0.69 |
ENSDART00000093199
ENSDART00000127519 ENSDART00000157142 |
tead3b
|
TEA domain family member 3 b |
| chr20_+_46620374 | 0.67 |
ENSDART00000005548
|
batf
|
basic leucine zipper transcription factor, ATF-like |
| chr24_+_36204028 | 0.66 |
ENSDART00000063832
ENSDART00000155260 |
rbbp8
|
retinoblastoma binding protein 8 |
| chr5_-_19006290 | 0.65 |
ENSDART00000137022
|
golga3
|
golgin A3 |
| chr20_-_37813863 | 0.65 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr7_-_12464412 | 0.64 |
ENSDART00000178723
|
adamtsl3
|
ADAMTS-like 3 |
| chr23_+_42254960 | 0.64 |
ENSDART00000102980
|
zcchc11
|
zinc finger, CCHC domain containing 11 |
| chr8_+_49117518 | 0.64 |
ENSDART00000079631
|
rad21l1
|
RAD21 cohesin complex component like 1 |
| chr23_-_900795 | 0.63 |
ENSDART00000190517
ENSDART00000182849 ENSDART00000111456 ENSDART00000185430 |
rbm10
|
RNA binding motif protein 10 |
| chr16_+_40043673 | 0.63 |
ENSDART00000102552
ENSDART00000125484 |
trmt11
|
tRNA methyltransferase 11 homolog (S. cerevisiae) |
| chr9_+_17983463 | 0.63 |
ENSDART00000182150
|
akap11
|
A kinase (PRKA) anchor protein 11 |
| chr17_-_41798856 | 0.63 |
ENSDART00000156031
ENSDART00000192801 ENSDART00000180172 ENSDART00000084745 ENSDART00000175577 |
ralgapa2
|
Ral GTPase activating protein, alpha subunit 2 (catalytic) |
| chr9_+_54695981 | 0.63 |
ENSDART00000183605
|
rab9a
|
RAB9A, member RAS oncogene family |
| chr15_+_29140126 | 0.63 |
ENSDART00000060034
|
zgc:113149
|
zgc:113149 |
| chr15_+_21262917 | 0.63 |
ENSDART00000101000
|
gkup
|
glucuronokinase with putative uridyl pyrophosphorylase |
| chr15_-_1038193 | 0.62 |
ENSDART00000159462
|
si:dkey-77f5.3
|
si:dkey-77f5.3 |
| chr19_+_4051535 | 0.62 |
ENSDART00000167084
|
btr24
|
bloodthirsty-related gene family, member 24 |
| chr6_-_40713183 | 0.60 |
ENSDART00000157113
ENSDART00000154810 ENSDART00000153702 |
si:ch211-157b11.12
|
si:ch211-157b11.12 |
| chr2_+_15048410 | 0.60 |
ENSDART00000058484
|
cnn3b
|
calponin 3, acidic b |
| chr6_-_12275836 | 0.60 |
ENSDART00000189980
|
pkp4
|
plakophilin 4 |
| chr7_-_59165640 | 0.59 |
ENSDART00000170853
|
haus6
|
HAUS augmin-like complex, subunit 6 |
| chr15_-_16177603 | 0.59 |
ENSDART00000156352
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| chr9_-_3934963 | 0.58 |
ENSDART00000062336
|
ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr18_-_43884044 | 0.58 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr11_-_23687158 | 0.57 |
ENSDART00000189599
|
pik3c2b
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr2_-_26720854 | 0.57 |
ENSDART00000148110
|
si:dkey-181m9.8
|
si:dkey-181m9.8 |
| chr6_-_24392426 | 0.57 |
ENSDART00000163965
|
brdt
|
bromodomain, testis-specific |
| chr10_+_2582254 | 0.57 |
ENSDART00000016103
|
nxnl2
|
nucleoredoxin like 2 |
| chr13_+_38817871 | 0.56 |
ENSDART00000187708
|
col19a1
|
collagen, type XIX, alpha 1 |
| chr23_-_31913231 | 0.56 |
ENSDART00000146852
ENSDART00000085054 |
mtfr2
|
mitochondrial fission regulator 2 |
| chr4_+_25917915 | 0.55 |
ENSDART00000138603
|
vezt
|
vezatin, adherens junctions transmembrane protein |
| chr2_-_33645411 | 0.55 |
ENSDART00000114663
|
b4galt2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr23_-_35790235 | 0.55 |
ENSDART00000142369
ENSDART00000141141 ENSDART00000011004 |
mfsd5
|
major facilitator superfamily domain containing 5 |
| chr8_-_22274222 | 0.54 |
ENSDART00000131805
|
nphp4
|
nephronophthisis 4 |
| chr10_-_2971407 | 0.53 |
ENSDART00000132526
|
marveld2a
|
MARVEL domain containing 2a |
| chr10_+_8197827 | 0.52 |
ENSDART00000026244
|
mtrex
|
Mtr4 exosome RNA helicase |
| chr8_-_21142550 | 0.51 |
ENSDART00000143192
ENSDART00000186820 ENSDART00000135938 |
cpt2
|
carnitine palmitoyltransferase 2 |
| chr20_-_28800999 | 0.51 |
ENSDART00000049462
|
rab15
|
RAB15, member RAS oncogene family |
| chr17_-_3815693 | 0.51 |
ENSDART00000161207
|
PLCB4
|
phospholipase C beta 4 |
| chr4_-_16124417 | 0.51 |
ENSDART00000128079
ENSDART00000077664 |
atp2b1a
|
ATPase plasma membrane Ca2+ transporting 1a |
| chr19_-_5103313 | 0.50 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr2_-_21819421 | 0.50 |
ENSDART00000121586
|
chd7
|
chromodomain helicase DNA binding protein 7 |
| chr22_+_16308450 | 0.50 |
ENSDART00000105678
|
lrrc39
|
leucine rich repeat containing 39 |
| chr17_+_37227936 | 0.50 |
ENSDART00000076009
|
hadhab
|
hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit alpha b |
| chr7_+_23292133 | 0.49 |
ENSDART00000134489
|
htr2cl1
|
5-hydroxytryptamine (serotonin) receptor 2C, G protein-coupled-like 1 |
| chr7_-_7764287 | 0.49 |
ENSDART00000173021
ENSDART00000113131 |
intu
|
inturned planar cell polarity protein |
| chr20_+_46513651 | 0.49 |
ENSDART00000152977
|
zc3h14
|
zinc finger CCCH-type containing 14 |
| chr2_-_39558643 | 0.49 |
ENSDART00000139860
ENSDART00000145231 ENSDART00000141721 |
cbln7
|
cerebellin 7 |
| chr17_+_12813316 | 0.49 |
ENSDART00000155629
|
ralgapa1
|
Ral GTPase activating protein, alpha subunit 1 (catalytic) |
| chr6_-_12172424 | 0.48 |
ENSDART00000109344
|
tanc1a
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1a |
| chr17_+_16046314 | 0.47 |
ENSDART00000154554
ENSDART00000154338 ENSDART00000155336 |
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr15_+_28410664 | 0.47 |
ENSDART00000132028
ENSDART00000057697 ENSDART00000057257 |
pitpnaa
|
phosphatidylinositol transfer protein, alpha a |
| chr19_-_20430892 | 0.47 |
ENSDART00000111409
|
tbc1d5
|
TBC1 domain family, member 5 |
| chr21_-_3853204 | 0.46 |
ENSDART00000188829
|
st6galnac4
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 |
| chr2_+_50608099 | 0.46 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr9_+_50001746 | 0.45 |
ENSDART00000058892
|
slc38a11
|
solute carrier family 38, member 11 |
| chr2_-_27619954 | 0.45 |
ENSDART00000144826
|
tgs1
|
trimethylguanosine synthase 1 |
| chr16_-_12491901 | 0.45 |
ENSDART00000174467
|
slc2a3b
|
solute carrier family 2 (facilitated glucose transporter), member 3b |
| chr9_-_43538328 | 0.45 |
ENSDART00000140526
|
znf385b
|
zinc finger protein 385B |
| chr2_-_32825917 | 0.45 |
ENSDART00000180409
|
prpf4ba
|
pre-mRNA processing factor 4Ba |
| chr12_+_30367079 | 0.44 |
ENSDART00000190112
|
ccdc186
|
si:ch211-225b10.4 |
| chr3_-_26805455 | 0.44 |
ENSDART00000180648
|
pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr8_-_52562672 | 0.44 |
ENSDART00000159333
ENSDART00000159974 |
si:ch73-199g24.2
|
si:ch73-199g24.2 |
| chr2_+_32826235 | 0.42 |
ENSDART00000143127
|
si:dkey-154p10.3
|
si:dkey-154p10.3 |
| chr12_-_11560794 | 0.42 |
ENSDART00000149098
ENSDART00000169975 |
plekha1b
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1b |
| chr9_-_5318873 | 0.41 |
ENSDART00000129308
|
ACVR1C
|
activin A receptor type 1C |
| chr20_+_29209767 | 0.41 |
ENSDART00000141252
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr10_+_35257651 | 0.40 |
ENSDART00000028940
|
styxl1
|
serine/threonine/tyrosine interacting-like 1 |
| chr2_-_28671139 | 0.39 |
ENSDART00000165272
ENSDART00000164657 |
dhcr7
|
7-dehydrocholesterol reductase |
| chr19_+_4968947 | 0.39 |
ENSDART00000003634
ENSDART00000134808 |
stard3
|
StAR-related lipid transfer (START) domain containing 3 |
| chr17_+_21902058 | 0.38 |
ENSDART00000142178
|
plekha1a
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1a |
| chr4_-_63769147 | 0.37 |
ENSDART00000189919
|
znf1098
|
zinc finger protein 1098 |
| chr24_-_34680956 | 0.37 |
ENSDART00000171009
|
ctnna1
|
catenin (cadherin-associated protein), alpha 1 |
| chr10_-_21955231 | 0.37 |
ENSDART00000183695
|
FO744833.2
|
|
| chr6_-_37744430 | 0.37 |
ENSDART00000150177
ENSDART00000149722 |
nipa2
|
non imprinted in Prader-Willi/Angelman syndrome 2 (human) |
| chr1_-_47071979 | 0.37 |
ENSDART00000160817
|
itsn1
|
intersectin 1 (SH3 domain protein) |
| chr17_-_48944465 | 0.37 |
ENSDART00000154110
|
si:ch1073-80i24.3
|
si:ch1073-80i24.3 |
| chr22_-_14696667 | 0.36 |
ENSDART00000180379
|
lrp1ba
|
low density lipoprotein receptor-related protein 1Ba |
| chr14_-_48961056 | 0.36 |
ENSDART00000124192
|
si:dkeyp-121d2.7
|
si:dkeyp-121d2.7 |
| chr4_-_73587205 | 0.36 |
ENSDART00000150837
|
znf1015
|
zinc finger protein 1015 |
| chr21_-_31013817 | 0.36 |
ENSDART00000065504
|
ncbp3
|
nuclear cap binding subunit 3 |
| chr8_+_52314542 | 0.36 |
ENSDART00000013059
ENSDART00000125241 |
dbnlb
|
drebrin-like b |
| chr2_+_6999369 | 0.36 |
ENSDART00000050597
|
xpr1b
|
xenotropic and polytropic retrovirus receptor 1b |
| chr2_-_32826108 | 0.36 |
ENSDART00000098834
|
prpf4ba
|
pre-mRNA processing factor 4Ba |
| chr11_-_29768054 | 0.35 |
ENSDART00000079117
|
plekha3
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 3 |
| chr5_-_8907819 | 0.34 |
ENSDART00000188523
|
adamts12
|
ADAM metallopeptidase with thrombospondin type 1 motif, 12 |
| chr20_-_29864390 | 0.34 |
ENSDART00000161834
ENSDART00000132278 |
rnf144ab
|
ring finger protein 144ab |
| chr3_-_26806032 | 0.34 |
ENSDART00000143710
|
pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr18_+_8833251 | 0.33 |
ENSDART00000143519
|
impdh1a
|
IMP (inosine 5'-monophosphate) dehydrogenase 1a |
| chr22_-_3564563 | 0.33 |
ENSDART00000145114
|
ptprsa
|
protein tyrosine phosphatase, receptor type, s, a |
| chr7_+_6915609 | 0.32 |
ENSDART00000159213
|
ccs
|
copper chaperone for superoxide dismutase |
| chr25_+_11456696 | 0.32 |
ENSDART00000171408
|
AGBL1
|
si:ch73-141f14.1 |
| chr24_-_41320037 | 0.32 |
ENSDART00000129058
|
rheb
|
Ras homolog, mTORC1 binding |
| chr11_-_2478374 | 0.32 |
ENSDART00000173205
|
si:ch73-267c23.10
|
si:ch73-267c23.10 |
| chr23_-_36446307 | 0.32 |
ENSDART00000136623
|
zgc:174906
|
zgc:174906 |
| chr20_+_29209926 | 0.31 |
ENSDART00000152949
ENSDART00000153016 |
katnbl1
|
katanin p80 subunit B-like 1 |
| chr19_-_27542433 | 0.31 |
ENSDART00000136414
|
si:ch211-152p11.4
|
si:ch211-152p11.4 |
| chr20_+_29209615 | 0.30 |
ENSDART00000062350
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr7_-_64589920 | 0.30 |
ENSDART00000172619
ENSDART00000184113 |
CR387919.1
|
|
| chr18_+_2228737 | 0.30 |
ENSDART00000165301
|
rab27a
|
RAB27A, member RAS oncogene family |
| chr15_+_44366556 | 0.30 |
ENSDART00000133449
|
GUCY1A2
|
guanylate cyclase 1 soluble subunit alpha 2 |
| chr2_+_56213694 | 0.30 |
ENSDART00000162582
|
upf1
|
upf1 regulator of nonsense transcripts homolog (yeast) |
| chr20_-_23226453 | 0.29 |
ENSDART00000142721
|
dcun1d4
|
DCN1, defective in cullin neddylation 1, domain containing 4 (S. cerevisiae) |
| chr18_-_15551360 | 0.28 |
ENSDART00000159915
ENSDART00000172690 |
ppfibp1b
|
PTPRF interacting protein, binding protein 1b (liprin beta 1) |
| chr20_+_13403281 | 0.27 |
ENSDART00000045183
ENSDART00000127350 |
enpp1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1 |
| chr4_-_2380173 | 0.27 |
ENSDART00000177727
|
nap1l1
|
nucleosome assembly protein 1-like 1 |
| chr23_+_4741543 | 0.27 |
ENSDART00000144761
|
raf1a
|
Raf-1 proto-oncogene, serine/threonine kinase a |
| chr16_-_40043322 | 0.27 |
ENSDART00000145278
|
si:dkey-29b11.3
|
si:dkey-29b11.3 |
| chr1_-_29139141 | 0.27 |
ENSDART00000075546
ENSDART00000133246 |
hsf2bp
|
heat shock transcription factor 2 binding protein |
| chr23_+_7548797 | 0.26 |
ENSDART00000006765
|
tm9sf4
|
transmembrane 9 superfamily protein member 4 |
| chr12_-_28349026 | 0.26 |
ENSDART00000183768
ENSDART00000152998 |
zgc:195081
|
zgc:195081 |
| chr22_-_881725 | 0.26 |
ENSDART00000035514
|
cept1b
|
choline/ethanolamine phosphotransferase 1b |
| chr9_+_1313418 | 0.26 |
ENSDART00000191361
|
casp8l2
|
caspase 8, apoptosis-related cysteine peptidase, like 2 |
| chr6_+_3280939 | 0.25 |
ENSDART00000151359
|
kdm4aa
|
lysine (K)-specific demethylase 4A, genome duplicate a |
| chr4_+_77943184 | 0.25 |
ENSDART00000159094
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr21_-_3770636 | 0.24 |
ENSDART00000053596
|
scamp1
|
secretory carrier membrane protein 1 |
| chr19_-_27542277 | 0.24 |
ENSDART00000114301
|
si:ch211-152p11.4
|
si:ch211-152p11.4 |
| chr11_+_39107131 | 0.24 |
ENSDART00000105140
|
zgc:112255
|
zgc:112255 |
| chr17_-_21162821 | 0.23 |
ENSDART00000157283
|
abhd12
|
abhydrolase domain containing 12 |
| chr17_+_12865746 | 0.23 |
ENSDART00000157083
|
ralgapa1
|
Ral GTPase activating protein, alpha subunit 1 (catalytic) |
| chr24_+_40860320 | 0.23 |
ENSDART00000161351
|
gorasp1b
|
golgi reassembly stacking protein 1b |
| chr11_+_33818179 | 0.23 |
ENSDART00000109418
|
spoplb
|
speckle-type POZ protein-like b |
| chr17_+_45454943 | 0.22 |
ENSDART00000074838
|
kcnk3b
|
potassium channel, subfamily K, member 3b |
| chr7_+_21841037 | 0.22 |
ENSDART00000077503
|
tm4sf5
|
transmembrane 4 L six family member 5 |
| chr3_-_16719244 | 0.21 |
ENSDART00000055859
|
pold1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr6_-_19665114 | 0.21 |
ENSDART00000168985
|
usp43a
|
ubiquitin specific peptidase 43a |
| chr13_+_31211050 | 0.21 |
ENSDART00000111705
|
ptpn20
|
protein tyrosine phosphatase, non-receptor type 20 |
| chr19_+_12406583 | 0.20 |
ENSDART00000013865
ENSDART00000151535 |
seh1l
|
SEH1-like (S. cerevisiae) |
| chr16_-_26855936 | 0.19 |
ENSDART00000167320
ENSDART00000078119 |
ino80c
|
INO80 complex subunit C |
| chr10_+_16036246 | 0.19 |
ENSDART00000141586
ENSDART00000135868 ENSDART00000065037 ENSDART00000124502 |
lmnb1
|
lamin B1 |
| chr16_+_5908483 | 0.18 |
ENSDART00000167393
|
ulk4
|
unc-51 like kinase 4 |
| chr19_+_9295244 | 0.18 |
ENSDART00000132255
ENSDART00000144299 |
si:ch73-15n24.1
|
si:ch73-15n24.1 |
| chr12_+_48803098 | 0.18 |
ENSDART00000074768
|
ppifb
|
peptidylprolyl isomerase Fb |
| chr7_+_7511914 | 0.17 |
ENSDART00000172848
|
clcn3
|
chloride channel 3 |
| chr19_-_205104 | 0.17 |
ENSDART00000011890
|
zbtb22a
|
zinc finger and BTB domain containing 22a |
Network of associatons between targets according to the STRING database.
First level regulatory network of meox1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.4 | 4.9 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.4 | 1.8 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.3 | 1.3 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.3 | 0.9 | GO:0021512 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.2 | 0.7 | GO:0042832 | response to protozoan(GO:0001562) defense response to protozoan(GO:0042832) |
| 0.2 | 0.7 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.2 | 1.8 | GO:0033206 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.2 | 0.6 | GO:0051039 | histone displacement(GO:0001207) regulation of transcription involved in meiotic cell cycle(GO:0051037) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.2 | 1.3 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.2 | 0.5 | GO:1900364 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.2 | 2.6 | GO:0060046 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.2 | 0.8 | GO:0010867 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.1 | 0.6 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 0.5 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 1.0 | GO:0016119 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.1 | 0.5 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.1 | 0.4 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.1 | 0.3 | GO:2000434 | regulation of protein neddylation(GO:2000434) |
| 0.1 | 1.5 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.1 | 0.9 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 1.3 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.1 | 0.8 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 2.8 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.1 | 0.4 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.1 | 0.4 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.1 | 0.6 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.1 | 1.2 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 1.1 | GO:0051445 | regulation of meiotic cell cycle(GO:0051445) |
| 0.1 | 0.7 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 0.3 | GO:0045598 | regulation of fat cell differentiation(GO:0045598) |
| 0.1 | 0.2 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 | 0.4 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.2 | GO:0052651 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.4 | GO:0014004 | microglia differentiation(GO:0014004) |
| 0.0 | 0.5 | GO:0048798 | rRNA transcription(GO:0009303) swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.0 | 0.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 1.1 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.2 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 | 0.6 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.3 | GO:0006177 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.0 | 0.1 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.0 | 0.3 | GO:1990748 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.8 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 1.8 | GO:0030217 | T cell differentiation(GO:0030217) |
| 0.0 | 1.4 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 0.8 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.2 | GO:0001765 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.3 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) synaptic membrane adhesion(GO:0099560) |
| 0.0 | 0.2 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.0 | 0.5 | GO:0060173 | limb development(GO:0060173) |
| 0.0 | 0.8 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.0 | 0.3 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.4 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.8 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.6 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.0 | 0.4 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.6 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.3 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 1.0 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 1.5 | GO:1902284 | axon extension involved in axon guidance(GO:0048846) neuron projection extension involved in neuron projection guidance(GO:1902284) |
| 0.0 | 0.4 | GO:0042446 | hormone biosynthetic process(GO:0042446) |
| 0.0 | 0.3 | GO:1903321 | negative regulation of protein ubiquitination(GO:0031397) negative regulation of protein modification by small protein conjugation or removal(GO:1903321) |
| 0.0 | 0.1 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.1 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.5 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 1.2 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.5 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 1.5 | GO:0017148 | negative regulation of translation(GO:0017148) |
| 0.0 | 0.1 | GO:0034164 | regulation of toll-like receptor 9 signaling pathway(GO:0034163) negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.0 | 0.3 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.4 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 1.2 | GO:0099518 | vesicle cytoskeletal trafficking(GO:0099518) |
| 0.0 | 0.0 | GO:0097237 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.0 | 0.3 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.2 | GO:0031060 | regulation of histone methylation(GO:0031060) |
| 0.0 | 0.3 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.3 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.0 | 0.3 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.3 | 0.9 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.3 | 4.9 | GO:0042555 | MCM complex(GO:0042555) |
| 0.2 | 0.8 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.1 | 0.6 | GO:1990498 | mitotic spindle microtubule(GO:1990498) |
| 0.1 | 0.5 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 0.8 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 1.0 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 0.6 | GO:0034991 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 0.6 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 0.6 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 0.5 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.7 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.4 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.2 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 2.0 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 1.1 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.7 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 0.9 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.5 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.4 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.5 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.7 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.1 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 6.1 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.5 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.2 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 1.3 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.4 | GO:0009374 | biotin binding(GO:0009374) |
| 0.5 | 2.4 | GO:1990518 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.4 | 2.0 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.3 | 1.3 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.2 | 0.7 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.2 | 0.5 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.2 | 0.6 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.2 | 2.6 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.8 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 2.6 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.4 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 0.9 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 1.0 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.1 | 0.7 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.1 | 1.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.2 | GO:0005252 | open rectifier potassium channel activity(GO:0005252) |
| 0.1 | 0.5 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 0.4 | GO:1902387 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.1 | 0.5 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 0.5 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 0.3 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 1.5 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 0.3 | GO:0004307 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 1.2 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 1.0 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 3.2 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 0.6 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 1.3 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.1 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.3 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 3.2 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.3 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 1.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.0 | 0.5 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.1 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.8 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.5 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.5 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 0.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.6 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 1.4 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.2 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.1 | GO:0004997 | thyrotropin-releasing hormone receptor activity(GO:0004997) |
| 0.0 | 0.3 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.0 | 0.1 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.6 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.3 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.3 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 1.3 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 1.0 | GO:0045296 | cadherin binding(GO:0045296) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 0.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.4 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.7 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.5 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 1.4 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.6 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.5 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.9 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 1.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 0.3 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 0.5 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.5 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.6 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 1.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.5 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.2 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.5 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 0.7 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.3 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 0.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.9 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |