Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
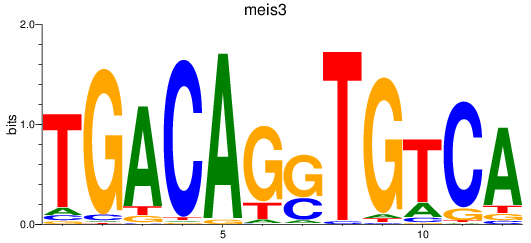
Results for meis3
Z-value: 1.27
Transcription factors associated with meis3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
meis3
|
ENSDARG00000002795 | myeloid ecotropic viral integration site 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| meis3 | dr11_v1_chr15_-_6863317_6863317 | 0.57 | 1.1e-01 | Click! |
Activity profile of meis3 motif
Sorted Z-values of meis3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_22310919 | 2.48 |
ENSDART00000108719
|
crygm2d10
|
crystallin, gamma M2d10 |
| chr16_-_16152199 | 2.33 |
ENSDART00000012718
|
fabp11b
|
fatty acid binding protein 11b |
| chr10_-_39011514 | 2.30 |
ENSDART00000075123
|
pcp4a
|
Purkinje cell protein 4a |
| chr9_-_22205682 | 2.22 |
ENSDART00000101869
|
crygm2d12
|
crystallin, gamma M2d12 |
| chr9_-_22232902 | 2.16 |
ENSDART00000101845
|
crygm2d5
|
crystallin, gamma M2d5 |
| chr9_-_22240052 | 2.10 |
ENSDART00000111109
|
crygm2d9
|
crystallin, gamma M2d9 |
| chr23_+_23658474 | 2.04 |
ENSDART00000162838
|
agrn
|
agrin |
| chr1_-_26026956 | 1.99 |
ENSDART00000102346
|
si:ch211-145b13.6
|
si:ch211-145b13.6 |
| chr6_-_14146979 | 1.98 |
ENSDART00000089564
|
cacnb4b
|
calcium channel, voltage-dependent, beta 4b subunit |
| chr1_-_40911332 | 1.91 |
ENSDART00000027463
|
hmx4
|
H6 family homeobox 4 |
| chr6_+_3827751 | 1.67 |
ENSDART00000003008
ENSDART00000122348 |
gad1b
|
glutamate decarboxylase 1b |
| chr11_-_41966854 | 1.60 |
ENSDART00000055709
|
her2
|
hairy-related 2 |
| chr17_+_7595356 | 1.55 |
ENSDART00000130625
|
si:dkeyp-110a12.4
|
si:dkeyp-110a12.4 |
| chr21_+_30351256 | 1.55 |
ENSDART00000078341
|
foxi3a
|
forkhead box I3a |
| chr7_-_26306546 | 1.52 |
ENSDART00000140817
|
zgc:77439
|
zgc:77439 |
| chr16_-_26537103 | 1.46 |
ENSDART00000134908
ENSDART00000008152 |
sgk2b
|
serum/glucocorticoid regulated kinase 2b |
| chr21_+_45733871 | 1.41 |
ENSDART00000187285
ENSDART00000193018 |
zgc:77058
|
zgc:77058 |
| chr2_-_32558795 | 1.40 |
ENSDART00000140026
|
smarcd3a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3a |
| chr15_-_29598679 | 1.40 |
ENSDART00000155153
|
si:ch211-207n23.2
|
si:ch211-207n23.2 |
| chr10_+_26667475 | 1.38 |
ENSDART00000133281
ENSDART00000147013 |
si:ch73-52f15.5
|
si:ch73-52f15.5 |
| chr8_+_8291492 | 1.35 |
ENSDART00000151314
|
srpk3
|
SRSF protein kinase 3 |
| chr5_-_46980651 | 1.29 |
ENSDART00000181022
ENSDART00000168038 |
edil3a
|
EGF-like repeats and discoidin I-like domains 3a |
| chr22_-_5006119 | 1.28 |
ENSDART00000187998
|
rx1
|
retinal homeobox gene 1 |
| chr2_+_6127593 | 1.23 |
ENSDART00000184007
|
fzr1b
|
fizzy/cell division cycle 20 related 1b |
| chr5_+_38913621 | 1.23 |
ENSDART00000137112
|
fras1
|
Fraser extracellular matrix complex subunit 1 |
| chr20_+_51104367 | 1.22 |
ENSDART00000073981
|
eif2s1b
|
eukaryotic translation initiation factor 2, subunit 1 alpha b |
| chr23_+_21663631 | 1.21 |
ENSDART00000066125
|
dhrs3a
|
dehydrogenase/reductase (SDR family) member 3a |
| chr11_-_17981421 | 1.16 |
ENSDART00000005999
ENSDART00000104046 |
twf2b
|
twinfilin actin-binding protein 2b |
| chr20_-_26001288 | 1.12 |
ENSDART00000136518
ENSDART00000063177 |
capn3b
|
calpain 3b |
| chr23_-_15216654 | 1.12 |
ENSDART00000131649
|
sulf2b
|
sulfatase 2b |
| chr16_-_7239457 | 1.08 |
ENSDART00000148992
ENSDART00000149260 |
nt5c3a
|
5'-nucleotidase, cytosolic IIIA |
| chr13_-_7031033 | 1.07 |
ENSDART00000193211
|
CABZ01061524.1
|
|
| chr2_+_45081489 | 1.06 |
ENSDART00000123966
|
chrng
|
cholinergic receptor, nicotinic, gamma |
| chr5_+_36752943 | 0.99 |
ENSDART00000017138
|
exoc3l2a
|
exocyst complex component 3-like 2a |
| chr22_-_34872533 | 0.98 |
ENSDART00000167176
|
slit1b
|
slit homolog 1b (Drosophila) |
| chr17_-_5583345 | 0.90 |
ENSDART00000035944
|
clic5a
|
chloride intracellular channel 5a |
| chr2_-_44183451 | 0.88 |
ENSDART00000111246
|
cadm3
|
cell adhesion molecule 3 |
| chr12_-_45876387 | 0.87 |
ENSDART00000043210
ENSDART00000149044 |
pax2b
|
paired box 2b |
| chr2_-_44183613 | 0.86 |
ENSDART00000079596
|
cadm3
|
cell adhesion molecule 3 |
| chr12_+_47122104 | 0.84 |
ENSDART00000184248
|
CABZ01088982.1
|
|
| chr5_+_37091626 | 0.83 |
ENSDART00000161054
|
tagln2
|
transgelin 2 |
| chr23_-_10177442 | 0.80 |
ENSDART00000144280
ENSDART00000129044 |
krt5
|
keratin 5 |
| chr2_+_47623202 | 0.77 |
ENSDART00000154465
|
si:ch211-165b10.3
|
si:ch211-165b10.3 |
| chr1_+_1805294 | 0.76 |
ENSDART00000103850
|
atp1a1a.3
|
ATPase Na+/K+ transporting subunit alpha 1a, tandem duplicate 3 |
| chr20_+_32406011 | 0.73 |
ENSDART00000018640
ENSDART00000137910 |
snx3
|
sorting nexin 3 |
| chr3_+_32557615 | 0.72 |
ENSDART00000151608
|
pax10
|
paired box 10 |
| chr7_+_65673885 | 0.70 |
ENSDART00000169182
|
parvab
|
parvin, alpha b |
| chr3_+_26734162 | 0.70 |
ENSDART00000114552
|
si:dkey-202l16.5
|
si:dkey-202l16.5 |
| chr1_+_34496855 | 0.69 |
ENSDART00000012873
|
klf12a
|
Kruppel-like factor 12a |
| chr7_-_67214972 | 0.68 |
ENSDART00000156861
|
swap70a
|
switching B cell complex subunit SWAP70a |
| chr10_+_406146 | 0.60 |
ENSDART00000145124
|
dact3a
|
dishevelled-binding antagonist of beta-catenin 3a |
| chr8_-_32506569 | 0.59 |
ENSDART00000061792
|
RFESD (1 of many)
|
Rieske Fe-S domain containing |
| chr9_-_25181137 | 0.58 |
ENSDART00000192624
|
esd
|
esterase D/formylglutathione hydrolase |
| chr16_-_1502699 | 0.55 |
ENSDART00000187189
|
sim1a
|
single-minded family bHLH transcription factor 1a |
| chr21_+_25231160 | 0.55 |
ENSDART00000063089
ENSDART00000139127 |
gng8
|
guanine nucleotide binding protein (G protein), gamma 8 |
| chr8_-_7444390 | 0.53 |
ENSDART00000149671
|
hdac6
|
histone deacetylase 6 |
| chr20_+_44311448 | 0.52 |
ENSDART00000114660
|
opn8b
|
opsin 8, group member b |
| chr7_-_30174882 | 0.50 |
ENSDART00000110409
|
frmd5
|
FERM domain containing 5 |
| chr5_+_23118470 | 0.50 |
ENSDART00000149893
|
nexmifa
|
neurite extension and migration factor a |
| chr5_+_24245682 | 0.50 |
ENSDART00000049003
|
atp6v1aa
|
ATPase H+ transporting V1 subunit Aa |
| chr25_-_22191983 | 0.49 |
ENSDART00000191181
|
pkp3a
|
plakophilin 3a |
| chr16_+_24971717 | 0.49 |
ENSDART00000156958
|
fpr1
|
formyl peptide receptor 1 |
| chr12_-_45875946 | 0.47 |
ENSDART00000149970
|
pax2b
|
paired box 2b |
| chr5_-_23118290 | 0.46 |
ENSDART00000132857
|
uprt
|
uracil phosphoribosyltransferase (FUR1) homolog (S. cerevisiae) |
| chr15_-_34056733 | 0.43 |
ENSDART00000170130
ENSDART00000188272 |
VWDE
|
si:dkey-30e9.7 |
| chr8_-_26609259 | 0.43 |
ENSDART00000027301
|
sema3b
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B |
| chr15_-_29598444 | 0.41 |
ENSDART00000154847
|
si:ch211-207n23.2
|
si:ch211-207n23.2 |
| chr19_-_35596207 | 0.41 |
ENSDART00000136811
|
col8a2
|
collagen, type VIII, alpha 2 |
| chr8_-_7444155 | 0.37 |
ENSDART00000148993
|
hdac6
|
histone deacetylase 6 |
| chr14_-_17305657 | 0.37 |
ENSDART00000168853
|
jakmip1
|
janus kinase and microtubule interacting protein 1 |
| chr5_-_21030934 | 0.34 |
ENSDART00000133461
ENSDART00000098667 |
camk2b1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 1 |
| chr25_+_32525131 | 0.34 |
ENSDART00000156145
|
scaper
|
S-phase cyclin A-associated protein in the ER |
| chr3_-_45308394 | 0.34 |
ENSDART00000155324
|
pdpk1a
|
3-phosphoinositide dependent protein kinase 1a |
| chr25_-_22191733 | 0.32 |
ENSDART00000067478
|
pkp3a
|
plakophilin 3a |
| chr6_+_33537267 | 0.32 |
ENSDART00000040334
|
pik3r3b
|
phosphoinositide-3-kinase, regulatory subunit 3b (gamma) |
| chr17_-_30839338 | 0.32 |
ENSDART00000139707
|
gdf7
|
growth differentiation factor 7 |
| chr9_-_28399071 | 0.26 |
ENSDART00000104317
ENSDART00000064343 |
klf7b
|
Kruppel-like factor 7b |
| chr17_-_15667198 | 0.26 |
ENSDART00000142972
ENSDART00000132571 ENSDART00000189936 |
manea
|
mannosidase, endo-alpha |
| chr19_-_30565122 | 0.26 |
ENSDART00000185650
|
hpcal4
|
hippocalcin like 4 |
| chr25_-_28600433 | 0.26 |
ENSDART00000138980
|
agbl2
|
ATP/GTP binding protein-like 2 |
| chr8_-_11229523 | 0.26 |
ENSDART00000002164
|
unc45b
|
unc-45 myosin chaperone B |
| chr1_-_10347198 | 0.25 |
ENSDART00000109141
ENSDART00000152565 |
si:ch211-243g6.3
|
si:ch211-243g6.3 |
| chr24_+_34806822 | 0.25 |
ENSDART00000148407
ENSDART00000188328 |
mchr2
|
melanin-concentrating hormone receptor 2 |
| chr12_+_36413886 | 0.24 |
ENSDART00000126325
|
si:ch211-250n8.1
|
si:ch211-250n8.1 |
| chr20_-_23171430 | 0.24 |
ENSDART00000109234
|
spata18
|
spermatogenesis associated 18 |
| chr1_-_44084071 | 0.23 |
ENSDART00000166912
|
vwa11
|
von Willebrand factor A domain containing 11 |
| chr22_+_30335936 | 0.20 |
ENSDART00000059923
|
mxi1
|
max interactor 1, dimerization protein |
| chr8_-_4508248 | 0.20 |
ENSDART00000141915
|
si:ch211-166a6.5
|
si:ch211-166a6.5 |
| chr2_+_9918678 | 0.20 |
ENSDART00000081253
|
acbd5b
|
acyl-CoA binding domain containing 5b |
| chr2_+_9918935 | 0.19 |
ENSDART00000140434
|
acbd5b
|
acyl-CoA binding domain containing 5b |
| chr24_-_30091937 | 0.18 |
ENSDART00000148249
|
plppr4b
|
phospholipid phosphatase related 4b |
| chr15_-_4415917 | 0.16 |
ENSDART00000062874
|
atp1b3b
|
ATPase Na+/K+ transporting subunit beta 3b |
| chr17_+_35362851 | 0.16 |
ENSDART00000137659
|
cmpk2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial |
| chr6_+_9107063 | 0.15 |
ENSDART00000083820
|
vps16
|
vacuolar protein sorting protein 16 |
| chr6_+_9952678 | 0.15 |
ENSDART00000019325
|
cyp20a1
|
cytochrome P450, family 20, subfamily A, polypeptide 1 |
| chr6_-_13408680 | 0.14 |
ENSDART00000151566
|
fmnl2b
|
formin-like 2b |
| chr18_+_36769758 | 0.13 |
ENSDART00000180375
ENSDART00000136463 ENSDART00000133487 ENSDART00000130206 |
fosb
|
FBJ murine osteosarcoma viral oncogene homolog B |
| chr25_-_3139805 | 0.10 |
ENSDART00000166625
|
epx
|
eosinophil peroxidase |
| chr19_+_24872159 | 0.09 |
ENSDART00000158490
|
si:ch211-195b13.1
|
si:ch211-195b13.1 |
| chr23_-_764135 | 0.09 |
ENSDART00000010248
ENSDART00000183978 |
mitfb
|
microphthalmia-associated transcription factor b |
| chr20_-_26467307 | 0.07 |
ENSDART00000078072
ENSDART00000158213 |
akap12b
|
A kinase (PRKA) anchor protein 12b |
| chr24_+_12989727 | 0.07 |
ENSDART00000126842
ENSDART00000129309 |
flj11011l
|
hypothetical protein FLJ11011-like (H. sapiens) |
| chr8_-_8698607 | 0.07 |
ENSDART00000046712
|
zgc:86609
|
zgc:86609 |
| chr2_+_15128418 | 0.05 |
ENSDART00000141921
|
arhgap29b
|
Rho GTPase activating protein 29b |
| chr14_-_26436951 | 0.05 |
ENSDART00000140173
|
si:dkeyp-110e4.6
|
si:dkeyp-110e4.6 |
| chr6_+_48041759 | 0.05 |
ENSDART00000140086
|
si:dkey-92f12.2
|
si:dkey-92f12.2 |
| chr8_-_27515540 | 0.05 |
ENSDART00000146132
|
si:ch211-254n4.3
|
si:ch211-254n4.3 |
| chr19_-_42588510 | 0.04 |
ENSDART00000102583
|
sytl1
|
synaptotagmin-like 1 |
| chr6_+_15762647 | 0.04 |
ENSDART00000127133
ENSDART00000128939 |
iqca1
|
IQ motif containing with AAA domain 1 |
| chr23_+_23182037 | 0.04 |
ENSDART00000137353
|
klhl17
|
kelch-like family member 17 |
| chr13_+_30169681 | 0.03 |
ENSDART00000138326
|
pald1b
|
phosphatase domain containing, paladin 1b |
| chr21_+_30194904 | 0.03 |
ENSDART00000137023
ENSDART00000078403 |
BRD8
|
si:ch211-59d17.3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of meis3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.6 | 1.7 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.4 | 1.5 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.4 | 1.1 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.3 | 1.6 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.2 | 1.2 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.2 | 1.2 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.2 | 2.0 | GO:0018120 | peptidyl-arginine ADP-ribosylation(GO:0018120) |
| 0.2 | 0.6 | GO:0072020 | proximal straight tubule development(GO:0072020) |
| 0.2 | 1.9 | GO:1900052 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.2 | 1.4 | GO:0031498 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.1 | 0.5 | GO:2000048 | negative regulation of cell-substrate adhesion(GO:0010812) negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.1 | 1.2 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.1 | 0.6 | GO:0046292 | formaldehyde metabolic process(GO:0046292) formaldehyde catabolic process(GO:0046294) |
| 0.1 | 0.5 | GO:0002283 | neutrophil activation involved in immune response(GO:0002283) |
| 0.1 | 1.1 | GO:0035860 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.1 | 1.4 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.1 | 9.2 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.1 | 0.3 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.1 | 1.1 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.1 | 0.2 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.0 | 2.0 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 1.2 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.0 | 1.2 | GO:0021602 | cranial nerve morphogenesis(GO:0021602) |
| 0.0 | 1.3 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 0.7 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 1.3 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.3 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.2 | GO:0042144 | vacuole fusion, non-autophagic(GO:0042144) |
| 0.0 | 1.5 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 0.7 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 1.6 | GO:1990266 | neutrophil migration(GO:1990266) |
| 0.0 | 0.9 | GO:0006476 | protein deacetylation(GO:0006476) |
| 0.0 | 0.5 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 1.2 | GO:0006413 | translational initiation(GO:0006413) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.1 | 1.1 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 1.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 0.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.6 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 1.2 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.8 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 1.0 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.6 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.2 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.5 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 2.0 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.9 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.2 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.9 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 1.6 | GO:0016324 | apical plasma membrane(GO:0016324) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.2 | 1.2 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.2 | 0.9 | GO:0047611 | tubulin deacetylase activity(GO:0042903) acetylspermidine deacetylase activity(GO:0047611) |
| 0.2 | 2.0 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.2 | 1.5 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.2 | 1.2 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 8.9 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 2.0 | GO:0043236 | laminin binding(GO:0043236) |
| 0.1 | 0.8 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 2.0 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 1.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 1.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.2 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 1.0 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.5 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 1.1 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 1.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.4 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.6 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.5 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 1.3 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.2 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.7 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.3 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.3 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 2.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.5 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.6 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.6 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.9 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.5 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.8 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 1.1 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.6 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.0 | 0.9 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |