Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
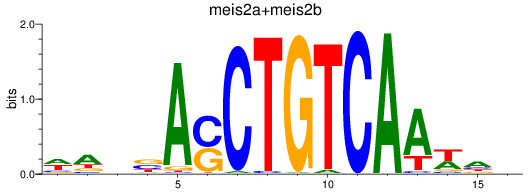
Results for meis2a+meis2b_FO834857.1_pknox1.1+pknox1.2
Z-value: 1.94
Transcription factors associated with meis2a+meis2b_FO834857.1_pknox1.1+pknox1.2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
meis2b
|
ENSDARG00000077840 | Meis homeobox 2a |
|
meis2a
|
ENSDARG00000098240 | Meis homeobox 2a |
|
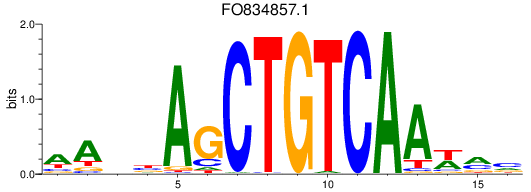
FO834857.1
|
ENSDARG00000112895 | homeobox protein TGIF2-like |
|
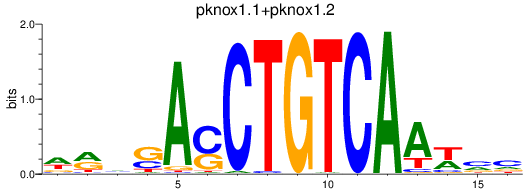
pknox1.1
|
ENSDARG00000018765 | pbx/knotted 1 homeobox 1.1 |
|
pknox1.2
|
ENSDARG00000036542 | pbx/knotted 1 homeobox 1.2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| meis2b | dr11_v1_chr20_-_10120442_10120442 | -0.90 | 1.1e-03 | Click! |
| FO834857.1 | dr11_v1_chr6_-_1814457_1814457 | 0.85 | 4.0e-03 | Click! |
| meis2a | dr11_v1_chr17_+_52822831_52822831 | -0.59 | 9.7e-02 | Click! |
| pknox1.1 | dr11_v1_chr9_+_19529951_19529951 | 0.53 | 1.4e-01 | Click! |
| pknox1.2 | dr11_v1_chr1_-_46984142_46984142 | -0.33 | 3.9e-01 | Click! |
Activity profile of meis2a+meis2b_FO834857.1_pknox1.1+pknox1.2 motif
Sorted Z-values of meis2a+meis2b_FO834857.1_pknox1.1+pknox1.2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_25959940 | 3.72 |
ENSDART00000143011
ENSDART00000140626 |
si:dkey-72l14.4
|
si:dkey-72l14.4 |
| chr17_-_36860988 | 3.29 |
ENSDART00000154981
|
senp6b
|
SUMO1/sentrin specific peptidase 6b |
| chr8_-_32385989 | 3.08 |
ENSDART00000143716
ENSDART00000098850 |
lipg
|
lipase, endothelial |
| chr14_-_41468892 | 2.96 |
ENSDART00000173099
ENSDART00000003170 |
mid1ip1l
|
MID1 interacting protein 1, like |
| chr12_-_48168135 | 2.77 |
ENSDART00000186624
|
pald1a
|
phosphatase domain containing, paladin 1a |
| chr14_-_6402769 | 2.77 |
ENSDART00000121552
|
slc44a1b
|
solute carrier family 44 (choline transporter), member 1b |
| chr6_+_49052741 | 2.66 |
ENSDART00000011876
|
sycp1
|
synaptonemal complex protein 1 |
| chr22_-_37834312 | 2.56 |
ENSDART00000076128
|
ppp1r2
|
protein phosphatase 1, regulatory (inhibitor) subunit 2 |
| chr3_+_7771420 | 2.55 |
ENSDART00000156809
ENSDART00000156309 |
hook2
|
hook microtubule-tethering protein 2 |
| chr16_+_50434668 | 2.53 |
ENSDART00000193500
|
IGLON5
|
zgc:110372 |
| chr23_-_18024543 | 2.52 |
ENSDART00000139695
|
pm20d1.1
|
peptidase M20 domain containing 1, tandem duplicate 1 |
| chr12_+_18916285 | 2.49 |
ENSDART00000127536
|
cbx7b
|
chromobox homolog 7b |
| chr17_-_7792376 | 2.49 |
ENSDART00000064655
|
zbtb2a
|
zinc finger and BTB domain containing 2a |
| chr2_-_11027258 | 2.40 |
ENSDART00000081072
ENSDART00000193824 ENSDART00000187036 ENSDART00000097741 |
ssbp3a
|
single stranded DNA binding protein 3a |
| chr20_-_28433990 | 2.38 |
ENSDART00000182824
ENSDART00000193381 |
wdr21
|
WD repeat domain 21 |
| chr10_+_37500234 | 2.36 |
ENSDART00000132096
ENSDART00000099473 |
msi2a
|
musashi RNA-binding protein 2a |
| chr9_+_27876146 | 2.35 |
ENSDART00000133997
|
armc8
|
armadillo repeat containing 8 |
| chr23_+_39963599 | 2.33 |
ENSDART00000166539
|
fyco1a
|
FYVE and coiled-coil domain containing 1a |
| chr18_+_35128685 | 2.31 |
ENSDART00000151579
|
si:ch211-195m9.3
|
si:ch211-195m9.3 |
| chr23_-_31060350 | 2.30 |
ENSDART00000145598
ENSDART00000191491 |
si:ch211-197l9.5
|
si:ch211-197l9.5 |
| chr11_+_44503774 | 2.29 |
ENSDART00000169295
|
ero1b
|
endoplasmic reticulum oxidoreductase beta |
| chr19_-_6134802 | 2.28 |
ENSDART00000140051
|
cica
|
capicua transcriptional repressor a |
| chr11_+_5880562 | 2.26 |
ENSDART00000129663
ENSDART00000130768 ENSDART00000160909 |
dazap1
|
DAZ associated protein 1 |
| chr4_+_27898833 | 2.25 |
ENSDART00000146099
|
cerk
|
ceramide kinase |
| chr16_-_51072406 | 2.17 |
ENSDART00000083777
|
ago3a
|
argonaute RISC catalytic component 3a |
| chr13_+_46803979 | 2.15 |
ENSDART00000159260
|
CU695232.1
|
|
| chr5_+_27137473 | 2.15 |
ENSDART00000181833
|
unc5db
|
unc-5 netrin receptor Db |
| chr4_-_20081621 | 2.11 |
ENSDART00000024647
|
dennd6b
|
DENN/MADD domain containing 6B |
| chr2_-_38117538 | 2.05 |
ENSDART00000013676
|
chd8
|
chromodomain helicase DNA binding protein 8 |
| chr23_-_36592436 | 2.05 |
ENSDART00000168246
|
spryd3
|
SPRY domain containing 3 |
| chr13_+_45524475 | 2.03 |
ENSDART00000074567
ENSDART00000019113 |
maco1b
|
macoilin 1b |
| chr6_-_57539141 | 2.02 |
ENSDART00000156967
|
itcha
|
itchy E3 ubiquitin protein ligase a |
| chr1_+_24382651 | 2.00 |
ENSDART00000123789
|
qdprb2
|
quinoid dihydropteridine reductase b2 |
| chr5_-_41124241 | 1.95 |
ENSDART00000083561
|
mtmr12
|
myotubularin related protein 12 |
| chr14_+_29200772 | 1.95 |
ENSDART00000166608
|
TENM2
|
si:dkey-34l15.2 |
| chr21_-_25612658 | 1.94 |
ENSDART00000115276
|
fibpb
|
fibroblast growth factor (acidic) intracellular binding protein b |
| chr13_+_23104134 | 1.94 |
ENSDART00000110266
|
pik3ap1
|
phosphoinositide-3-kinase adaptor protein 1 |
| chr6_-_41135215 | 1.93 |
ENSDART00000001861
|
slc6a22.1
|
solute carrier family 6 member 22, tandem duplicate 1 |
| chr8_-_41519234 | 1.92 |
ENSDART00000167283
ENSDART00000180666 |
golga1
|
golgin A1 |
| chr5_-_69482891 | 1.91 |
ENSDART00000109487
|
CABZ01032476.1
|
|
| chr22_-_16377960 | 1.90 |
ENSDART00000168170
|
ttc39c
|
tetratricopeptide repeat domain 39C |
| chr14_-_34605607 | 1.90 |
ENSDART00000191608
|
afap1l1a
|
actin filament associated protein 1-like 1a |
| chr19_+_42231431 | 1.87 |
ENSDART00000102698
|
jtb
|
jumping translocation breakpoint |
| chr15_-_28587147 | 1.86 |
ENSDART00000156049
|
ssh2a
|
slingshot protein phosphatase 2a |
| chr13_-_24396003 | 1.84 |
ENSDART00000016211
|
tbp
|
TATA box binding protein |
| chr11_-_34478225 | 1.83 |
ENSDART00000189604
|
xxylt1
|
xyloside xylosyltransferase 1 |
| chr7_+_16352924 | 1.82 |
ENSDART00000158972
|
mpped2a
|
metallophosphoesterase domain containing 2a |
| chr25_-_1323623 | 1.80 |
ENSDART00000156532
ENSDART00000157163 ENSDART00000156062 ENSDART00000082447 ENSDART00000189175 |
calml4b
|
calmodulin-like 4b |
| chr15_+_23528310 | 1.79 |
ENSDART00000152523
|
si:dkey-182i3.8
|
si:dkey-182i3.8 |
| chr22_-_16377666 | 1.78 |
ENSDART00000161878
|
ttc39c
|
tetratricopeptide repeat domain 39C |
| chr8_-_41519064 | 1.78 |
ENSDART00000098578
ENSDART00000112214 |
golga1
|
golgin A1 |
| chr19_-_46032556 | 1.77 |
ENSDART00000167185
|
nup153
|
nucleoporin 153 |
| chr14_+_30340251 | 1.77 |
ENSDART00000148448
|
mtus1a
|
microtubule associated tumor suppressor 1a |
| chr14_-_34605804 | 1.76 |
ENSDART00000144547
|
afap1l1a
|
actin filament associated protein 1-like 1a |
| chr15_-_506010 | 1.69 |
ENSDART00000155472
|
nudt8
|
nudix (nucleoside diphosphate linked moiety X)-type motif 8 |
| chr15_-_28587490 | 1.68 |
ENSDART00000186196
|
ssh2a
|
slingshot protein phosphatase 2a |
| chr8_-_410728 | 1.67 |
ENSDART00000151255
|
trim36
|
tripartite motif containing 36 |
| chr10_-_35177257 | 1.66 |
ENSDART00000077426
|
pom121
|
POM121 transmembrane nucleoporin |
| chr20_+_1329509 | 1.64 |
ENSDART00000017791
ENSDART00000136669 |
tab2
|
TGF-beta activated kinase 1/MAP3K7 binding protein 2 |
| chr17_+_33999630 | 1.63 |
ENSDART00000167085
ENSDART00000155030 ENSDART00000168522 ENSDART00000191799 ENSDART00000189684 ENSDART00000153942 ENSDART00000187272 ENSDART00000127692 |
gphna
|
gephyrin a |
| chr5_-_13766651 | 1.62 |
ENSDART00000134064
|
mxd1
|
MAX dimerization protein 1 |
| chr7_-_37555208 | 1.61 |
ENSDART00000148905
ENSDART00000150229 |
cylda
|
cylindromatosis (turban tumor syndrome), a |
| chr15_+_23528010 | 1.61 |
ENSDART00000152786
|
si:dkey-182i3.8
|
si:dkey-182i3.8 |
| chr19_+_43905671 | 1.60 |
ENSDART00000168725
ENSDART00000133628 |
ankib1a
|
ankyrin repeat and IBR domain containing 1a |
| chr21_+_39336285 | 1.58 |
ENSDART00000139677
|
si:ch211-274p24.4
|
si:ch211-274p24.4 |
| chr23_+_13528053 | 1.57 |
ENSDART00000162217
|
uckl1b
|
uridine-cytidine kinase 1-like 1b |
| chr12_-_33659328 | 1.57 |
ENSDART00000153457
|
tmem94
|
transmembrane protein 94 |
| chr24_-_41180149 | 1.56 |
ENSDART00000019975
|
acvr2ba
|
activin A receptor type 2Ba |
| chr6_-_10809546 | 1.55 |
ENSDART00000151661
|
wipf1b
|
WAS/WASL interacting protein family, member 1b |
| chr13_-_24396199 | 1.54 |
ENSDART00000181093
|
tbp
|
TATA box binding protein |
| chr9_-_12575776 | 1.53 |
ENSDART00000128931
ENSDART00000182695 |
igf2bp2a
|
insulin-like growth factor 2 mRNA binding protein 2a |
| chr20_+_3339248 | 1.52 |
ENSDART00000108955
|
mfsd4b
|
major facilitator superfamily domain containing 4B |
| chr25_+_14870043 | 1.52 |
ENSDART00000035714
ENSDART00000171835 |
dnajc24
|
DnaJ (Hsp40) homolog, subfamily C, member 24 |
| chr18_+_39487486 | 1.52 |
ENSDART00000126978
|
acadl
|
acyl-CoA dehydrogenase long chain |
| chr17_+_19630068 | 1.47 |
ENSDART00000182619
|
rgs7a
|
regulator of G protein signaling 7a |
| chr3_+_29082267 | 1.46 |
ENSDART00000145615
|
cacna1i
|
calcium channel, voltage-dependent, T type, alpha 1I subunit |
| chr18_+_907266 | 1.44 |
ENSDART00000171729
|
pkma
|
pyruvate kinase M1/2a |
| chr1_+_45217425 | 1.44 |
ENSDART00000179983
ENSDART00000074683 |
EVI5L
|
si:ch211-239f4.1 |
| chr5_-_18474486 | 1.44 |
ENSDART00000090580
|
si:dkey-215k6.1
|
si:dkey-215k6.1 |
| chr7_+_52761841 | 1.41 |
ENSDART00000111444
|
ppip5k1a
|
diphosphoinositol pentakisphosphate kinase 1a |
| chr24_-_36095526 | 1.41 |
ENSDART00000158145
|
CABZ01075509.1
|
|
| chr9_-_52490579 | 1.40 |
ENSDART00000161667
|
smarcal1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a-like 1 |
| chr21_-_14832369 | 1.39 |
ENSDART00000144859
|
pus1
|
pseudouridylate synthase 1 |
| chr2_+_30489846 | 1.38 |
ENSDART00000145732
|
march6
|
membrane-associated ring finger (C3HC4) 6 |
| chr8_+_44623540 | 1.38 |
ENSDART00000141513
|
grk5l
|
G protein-coupled receptor kinase 5 like |
| chr7_-_9873652 | 1.38 |
ENSDART00000006343
|
asb7
|
ankyrin repeat and SOCS box containing 7 |
| chr17_+_50074372 | 1.36 |
ENSDART00000113644
|
vps39
|
vacuolar protein sorting 39 homolog (S. cerevisiae) |
| chr1_+_45217619 | 1.36 |
ENSDART00000125037
|
EVI5L
|
si:ch211-239f4.1 |
| chr2_-_45510699 | 1.35 |
ENSDART00000024034
ENSDART00000145634 |
gpsm2
|
G protein signaling modulator 2 |
| chr17_-_50040927 | 1.35 |
ENSDART00000184304
|
FO834825.1
|
|
| chr5_-_35200590 | 1.34 |
ENSDART00000051271
|
fcho2
|
FCH domain only 2 |
| chr6_+_38773376 | 1.33 |
ENSDART00000078128
ENSDART00000184053 |
ube3a
|
ubiquitin protein ligase E3A |
| chr13_-_7233811 | 1.32 |
ENSDART00000162026
|
ninl
|
ninein-like |
| chr23_+_21492151 | 1.31 |
ENSDART00000025487
|
icmt
|
isoprenylcysteine carboxyl methyltransferase |
| chr5_-_39805620 | 1.30 |
ENSDART00000137801
|
rasgef1ba
|
RasGEF domain family, member 1Ba |
| chr4_-_73394385 | 1.30 |
ENSDART00000174205
ENSDART00000143417 |
zgc:162958
|
zgc:162958 |
| chr2_+_27855346 | 1.29 |
ENSDART00000175159
ENSDART00000192645 |
buc
|
bucky ball |
| chr25_-_3192405 | 1.29 |
ENSDART00000104835
|
hps5
|
Hermansky-Pudlak syndrome 5 |
| chr7_-_56766100 | 1.29 |
ENSDART00000189934
|
csnk2a2a
|
casein kinase 2, alpha prime polypeptide a |
| chr21_-_13123176 | 1.25 |
ENSDART00000144866
ENSDART00000024616 |
fam219aa
|
family with sequence similarity 219, member Aa |
| chr9_+_33267211 | 1.24 |
ENSDART00000025635
|
usp9
|
ubiquitin specific peptidase 9 |
| chr9_-_12575569 | 1.24 |
ENSDART00000102419
|
igf2bp2a
|
insulin-like growth factor 2 mRNA binding protein 2a |
| chr25_+_22319940 | 1.24 |
ENSDART00000154065
ENSDART00000153492 ENSDART00000024866 ENSDART00000154376 |
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr20_-_20270191 | 1.24 |
ENSDART00000009356
|
ppp2r5ea
|
protein phosphatase 2, regulatory subunit B', epsilon isoform a |
| chr10_+_28160265 | 1.23 |
ENSDART00000022484
|
rnft1
|
ring finger protein, transmembrane 1 |
| chr5_-_23485161 | 1.23 |
ENSDART00000170293
ENSDART00000134069 |
si:dkeyp-20g2.1
si:dkeyp-20g2.3
|
si:dkeyp-20g2.1 si:dkeyp-20g2.3 |
| chr14_-_21238046 | 1.23 |
ENSDART00000129743
|
si:ch211-175m2.5
|
si:ch211-175m2.5 |
| chr14_-_33481428 | 1.23 |
ENSDART00000147059
ENSDART00000140001 ENSDART00000124242 ENSDART00000164836 ENSDART00000190104 ENSDART00000186833 ENSDART00000180873 |
lamp2
|
lysosomal-associated membrane protein 2 |
| chr22_+_1006573 | 1.23 |
ENSDART00000123458
|
pparda
|
peroxisome proliferator-activated receptor delta a |
| chr3_+_17030665 | 1.22 |
ENSDART00000159849
ENSDART00000174491 ENSDART00000104519 ENSDART00000080854 |
stat3
|
signal transducer and activator of transcription 3 (acute-phase response factor) |
| chr20_-_28433616 | 1.22 |
ENSDART00000169289
|
wdr21
|
WD repeat domain 21 |
| chr5_-_23675222 | 1.22 |
ENSDART00000135153
|
TBC1D8B
|
si:dkey-110k5.6 |
| chr14_+_23184517 | 1.21 |
ENSDART00000181410
|
enox2
|
ecto-NOX disulfide-thiol exchanger 2 |
| chr12_-_48467733 | 1.20 |
ENSDART00000153126
ENSDART00000152895 ENSDART00000014190 |
sec31b
|
SEC31 homolog B, COPII coat complex component |
| chr17_-_14780578 | 1.19 |
ENSDART00000154690
|
si:ch211-266o15.1
|
si:ch211-266o15.1 |
| chr3_+_13190012 | 1.19 |
ENSDART00000179747
ENSDART00000109876 ENSDART00000124824 ENSDART00000130261 |
sun1
|
Sad1 and UNC84 domain containing 1 |
| chr12_-_22039350 | 1.18 |
ENSDART00000153187
|
thrab
|
thyroid hormone receptor alpha b |
| chr20_+_27713210 | 1.18 |
ENSDART00000132222
|
zbtb1
|
zinc finger and BTB domain containing 1 |
| chr5_-_12587053 | 1.17 |
ENSDART00000162780
|
vsig10
|
V-set and immunoglobulin domain containing 10 |
| chr1_+_2301961 | 1.16 |
ENSDART00000108919
ENSDART00000143361 ENSDART00000142944 |
farp1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr4_-_18281070 | 1.16 |
ENSDART00000135330
ENSDART00000179075 ENSDART00000046871 |
uhrf1bp1l
|
UHRF1 binding protein 1-like |
| chr3_-_20793655 | 1.16 |
ENSDART00000163473
ENSDART00000159457 |
spop
|
speckle-type POZ protein |
| chr12_+_33320884 | 1.15 |
ENSDART00000188988
|
csnk1db
|
casein kinase 1, delta b |
| chr5_-_323712 | 1.15 |
ENSDART00000188793
|
HOOK3
|
hook microtubule tethering protein 3 |
| chr24_+_37484661 | 1.15 |
ENSDART00000165125
ENSDART00000109221 |
wdr90
|
WD repeat domain 90 |
| chr25_-_8138122 | 1.14 |
ENSDART00000104659
|
sergef
|
secretion regulating guanine nucleotide exchange factor |
| chr4_-_73562122 | 1.14 |
ENSDART00000174096
|
CU570689.2
|
|
| chr4_-_73561848 | 1.13 |
ENSDART00000174210
|
CU570689.2
|
|
| chr5_-_20135679 | 1.13 |
ENSDART00000079402
|
usp30
|
ubiquitin specific peptidase 30 |
| chr20_+_13783040 | 1.12 |
ENSDART00000115329
ENSDART00000152497 |
lpgat1
|
lysophosphatidylglycerol acyltransferase 1 |
| chr21_-_588858 | 1.12 |
ENSDART00000168983
|
TMEM38B
|
transmembrane protein 38B |
| chr7_+_14005111 | 1.10 |
ENSDART00000187365
|
furina
|
furin (paired basic amino acid cleaving enzyme) a |
| chr21_+_36623162 | 1.10 |
ENSDART00000027459
|
grk6
|
G protein-coupled receptor kinase 6 |
| chr13_+_13945218 | 1.10 |
ENSDART00000089501
ENSDART00000142997 |
eif2ak3
|
eukaryotic translation initiation factor 2-alpha kinase 3 |
| chr25_-_21894317 | 1.10 |
ENSDART00000089642
|
fbxo31
|
F-box protein 31 |
| chr22_-_21897203 | 1.10 |
ENSDART00000158501
ENSDART00000105566 ENSDART00000136795 |
gna11a
|
guanine nucleotide binding protein (G protein), alpha 11a (Gq class) |
| chr20_+_27712714 | 1.09 |
ENSDART00000008306
|
zbtb1
|
zinc finger and BTB domain containing 1 |
| chr23_-_19103855 | 1.09 |
ENSDART00000016901
|
pard6b
|
par-6 partitioning defective 6 homolog beta (C. elegans) |
| chr20_+_47953047 | 1.07 |
ENSDART00000079734
|
hadhaa
|
hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit alpha a |
| chr16_+_48714048 | 1.07 |
ENSDART00000148709
ENSDART00000150121 |
brd2b
|
bromodomain containing 2b |
| chr16_+_20167811 | 1.06 |
ENSDART00000004031
|
hacl1
|
2-hydroxyacyl-CoA lyase 1 |
| chr22_+_18316144 | 1.06 |
ENSDART00000137985
|
gatad2ab
|
GATA zinc finger domain containing 2Ab |
| chr6_+_33537267 | 1.06 |
ENSDART00000040334
|
pik3r3b
|
phosphoinositide-3-kinase, regulatory subunit 3b (gamma) |
| chr21_-_18262287 | 1.06 |
ENSDART00000176716
|
vav2
|
vav 2 guanine nucleotide exchange factor |
| chr3_-_23596809 | 1.06 |
ENSDART00000156897
|
ube2z
|
ubiquitin-conjugating enzyme E2Z |
| chr18_-_41160975 | 1.05 |
ENSDART00000187517
|
CABZ01005876.1
|
|
| chr13_+_24396666 | 1.05 |
ENSDART00000139197
ENSDART00000101200 |
zgc:153169
|
zgc:153169 |
| chr5_-_37103487 | 1.04 |
ENSDART00000149211
|
il13ra2
|
interleukin 13 receptor, alpha 2 |
| chr15_-_20125331 | 1.04 |
ENSDART00000152355
|
med13b
|
mediator complex subunit 13b |
| chr20_-_26391958 | 1.02 |
ENSDART00000078062
|
armt1
|
acidic residue methyltransferase 1 |
| chr22_-_37796998 | 1.01 |
ENSDART00000124742
ENSDART00000191232 |
acap2
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 |
| chr5_-_27993972 | 1.01 |
ENSDART00000175819
|
ppp3cca
|
protein phosphatase 3, catalytic subunit, gamma isozyme, a |
| chr14_-_38889840 | 1.00 |
ENSDART00000035779
|
zgc:101583
|
zgc:101583 |
| chr2_+_52232630 | 0.99 |
ENSDART00000006216
|
plpp2a
|
phospholipid phosphatase 2a |
| chr4_+_15006217 | 0.99 |
ENSDART00000090837
|
zc3hc1
|
zinc finger, C3HC-type containing 1 |
| chr5_-_32489796 | 0.98 |
ENSDART00000168870
|
gpr107
|
G protein-coupled receptor 107 |
| chr7_+_1534820 | 0.97 |
ENSDART00000192997
|
tox4b
|
TOX high mobility group box family member 4 b |
| chr25_-_2723682 | 0.97 |
ENSDART00000113382
|
adpgk
|
ADP-dependent glucokinase |
| chr20_+_21391181 | 0.97 |
ENSDART00000185158
ENSDART00000049586 ENSDART00000024922 |
jag2b
|
jagged 2b |
| chr11_+_28218141 | 0.96 |
ENSDART00000043756
|
ephb2b
|
eph receptor B2b |
| chr13_+_9612395 | 0.96 |
ENSDART00000136689
ENSDART00000138362 |
slf2
|
SMC5-SMC6 complex localization factor 2 |
| chr19_-_18313303 | 0.95 |
ENSDART00000164644
ENSDART00000167480 ENSDART00000163104 |
si:dkey-208k4.2
|
si:dkey-208k4.2 |
| chr4_-_27897160 | 0.95 |
ENSDART00000066924
ENSDART00000066925 ENSDART00000193020 |
tbc1d22a
|
TBC1 domain family, member 22a |
| chr14_-_33945692 | 0.95 |
ENSDART00000168546
ENSDART00000189778 |
zdhhc24
|
zinc finger, DHHC-type containing 24 |
| chr5_-_3991655 | 0.94 |
ENSDART00000159368
|
myo19
|
myosin XIX |
| chr16_-_42770064 | 0.94 |
ENSDART00000112879
|
slc4a7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr10_-_25328814 | 0.94 |
ENSDART00000123820
|
tmem135
|
transmembrane protein 135 |
| chr5_-_8614217 | 0.93 |
ENSDART00000164844
|
rictora
|
RPTOR independent companion of MTOR, complex 2 a |
| chr1_+_41666611 | 0.93 |
ENSDART00000145789
|
fbxo41
|
F-box protein 41 |
| chr16_-_35329803 | 0.92 |
ENSDART00000161729
ENSDART00000157700 ENSDART00000184584 ENSDART00000174713 ENSDART00000162518 |
ptprub
|
protein tyrosine phosphatase, receptor type, U, b |
| chr3_-_40836081 | 0.92 |
ENSDART00000143135
|
wipi2
|
WD repeat domain, phosphoinositide interacting 2 |
| chr17_+_38030327 | 0.91 |
ENSDART00000085481
|
slc25a21
|
solute carrier family 25 (mitochondrial oxoadipate carrier), member 21 |
| chr21_-_43474012 | 0.91 |
ENSDART00000065104
|
tmem185
|
transmembrane protein 185 |
| chr2_-_27651674 | 0.90 |
ENSDART00000177402
|
tgs1
|
trimethylguanosine synthase 1 |
| chr3_-_27880229 | 0.90 |
ENSDART00000151404
|
abat
|
4-aminobutyrate aminotransferase |
| chr12_-_10505986 | 0.90 |
ENSDART00000152672
|
zgc:152977
|
zgc:152977 |
| chr5_-_39805874 | 0.89 |
ENSDART00000176202
ENSDART00000191683 |
rasgef1ba
|
RasGEF domain family, member 1Ba |
| chr9_+_17984358 | 0.87 |
ENSDART00000192399
|
akap11
|
A kinase (PRKA) anchor protein 11 |
| chr11_+_42726712 | 0.87 |
ENSDART00000028955
|
tdrd3
|
tudor domain containing 3 |
| chr16_-_54978981 | 0.87 |
ENSDART00000154023
|
wdtc1
|
WD and tetratricopeptide repeats 1 |
| chr20_-_165818 | 0.86 |
ENSDART00000123860
|
si:ch211-241j12.3
|
si:ch211-241j12.3 |
| chr7_+_71776905 | 0.85 |
ENSDART00000189745
ENSDART00000161344 ENSDART00000158782 |
nsun6
|
NOL1/NOP2/Sun domain family, member 6 |
| chr2_-_42843797 | 0.84 |
ENSDART00000137913
|
adcy8
|
adenylate cyclase 8 (brain) |
| chr6_+_4371137 | 0.84 |
ENSDART00000168973
|
rbm26
|
RNA binding motif protein 26 |
| chr16_-_22775480 | 0.84 |
ENSDART00000141778
ENSDART00000145585 ENSDART00000125963 ENSDART00000127570 |
pbxip1b
|
pre-B-cell leukemia homeobox interacting protein 1b |
| chr7_+_55149001 | 0.84 |
ENSDART00000148642
|
cdh31
|
cadherin 31 |
| chr9_-_25366541 | 0.83 |
ENSDART00000021672
|
epc2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr16_+_11151699 | 0.83 |
ENSDART00000140674
|
cicb
|
capicua transcriptional repressor b |
| chr11_+_31680513 | 0.82 |
ENSDART00000139900
ENSDART00000040305 |
diaph3
|
diaphanous-related formin 3 |
| chr10_+_20590190 | 0.82 |
ENSDART00000131819
|
letm2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr9_+_29548630 | 0.80 |
ENSDART00000132295
|
rnf17
|
ring finger protein 17 |
| chr7_+_41887429 | 0.80 |
ENSDART00000115090
|
gpt2
|
glutamic pyruvate transaminase (alanine aminotransferase) 2 |
| chr3_+_17616201 | 0.80 |
ENSDART00000156775
|
rab5c
|
RAB5C, member RAS oncogene family |
| chr11_-_843811 | 0.79 |
ENSDART00000173331
|
atg7
|
ATG7 autophagy related 7 homolog (S. cerevisiae) |
| chr14_-_3944017 | 0.78 |
ENSDART00000055817
|
si:ch73-49o8.1
|
si:ch73-49o8.1 |
| chr1_+_18863060 | 0.78 |
ENSDART00000139241
|
rnf38
|
ring finger protein 38 |
| chr1_+_29183962 | 0.78 |
ENSDART00000113735
|
cars2
|
cysteinyl-tRNA synthetase 2, mitochondrial |
| chr8_-_13678415 | 0.78 |
ENSDART00000134153
ENSDART00000143331 |
si:dkey-258f14.3
|
si:dkey-258f14.3 |
| chr11_+_45436703 | 0.77 |
ENSDART00000168295
ENSDART00000173293 |
sos1
|
son of sevenless homolog 1 (Drosophila) |
| chr21_-_23046606 | 0.76 |
ENSDART00000016167
|
zw10
|
zw10 kinetochore protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of meis2a+meis2b_FO834857.1_pknox1.1+pknox1.2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:0051026 | meiotic DNA repair synthesis(GO:0000711) chiasma assembly(GO:0051026) |
| 0.8 | 3.3 | GO:0090230 | regulation of centromere complex assembly(GO:0090230) regulation of kinetochore assembly(GO:0090234) |
| 0.6 | 2.5 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.5 | 1.6 | GO:0072579 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.5 | 2.3 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.4 | 1.2 | GO:0044320 | leptin-mediated signaling pathway(GO:0033210) cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 0.4 | 2.0 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.4 | 2.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.3 | 1.5 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.3 | 2.1 | GO:2000178 | negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.3 | 1.1 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.3 | 1.4 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.3 | 1.6 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.3 | 1.3 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.3 | 2.8 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.2 | 1.2 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.2 | 1.2 | GO:1904292 | regulation of ERAD pathway(GO:1904292) |
| 0.2 | 1.2 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.2 | 1.1 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.2 | 3.7 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.2 | 0.9 | GO:0045922 | negative regulation of fatty acid metabolic process(GO:0045922) |
| 0.2 | 1.3 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.2 | 3.5 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.2 | 1.4 | GO:0048478 | embryonic body morphogenesis(GO:0010172) replication fork protection(GO:0048478) |
| 0.2 | 2.0 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.2 | 1.8 | GO:0010888 | negative regulation of cholesterol storage(GO:0010887) negative regulation of lipid storage(GO:0010888) |
| 0.2 | 0.8 | GO:0021543 | substrate-dependent cell migration(GO:0006929) pallium development(GO:0021543) cerebral cortex cell migration(GO:0021795) cerebral cortex tangential migration(GO:0021800) cerebral cortex tangential migration using cell-axon interactions(GO:0021824) substrate-dependent cerebral cortex tangential migration(GO:0021825) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) cerebral cortex development(GO:0021987) |
| 0.2 | 1.9 | GO:0072091 | regulation of stem cell proliferation(GO:0072091) |
| 0.2 | 1.3 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.2 | 1.3 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.2 | 0.9 | GO:0009450 | gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.2 | 1.4 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.2 | 1.8 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.2 | 0.7 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.2 | 1.4 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.2 | 1.9 | GO:0018410 | C-terminal protein amino acid modification(GO:0018410) |
| 0.2 | 0.7 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.2 | 0.5 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.2 | 1.1 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 0.6 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.1 | 1.5 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.1 | 2.5 | GO:0071166 | ribonucleoprotein complex localization(GO:0071166) |
| 0.1 | 0.4 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.1 | 2.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 1.1 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 1.2 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 0.8 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.5 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.1 | 0.7 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.1 | 1.0 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.1 | 0.5 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.1 | 0.6 | GO:0006535 | cysteine biosynthetic process from serine(GO:0006535) hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.1 | 0.9 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.1 | 1.2 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 0.3 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.1 | 0.5 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.1 | 3.4 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 1.4 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 0.4 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 1.4 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 3.0 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.1 | 0.7 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 0.7 | GO:0097107 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.1 | 2.6 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.1 | 1.5 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 0.5 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.1 | 3.3 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.1 | 0.5 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.1 | 0.6 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.1 | 0.5 | GO:0003272 | endocardial cushion formation(GO:0003272) |
| 0.1 | 0.3 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.1 | 0.9 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.2 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.1 | 1.0 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 1.1 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.1 | 0.9 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 | 1.1 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.1 | 0.4 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 0.7 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.1 | 0.2 | GO:0045040 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.1 | 0.3 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
| 0.1 | 0.3 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.1 | 0.3 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 0.1 | 0.3 | GO:0015682 | ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.1 | 0.8 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 0.3 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) protein insertion into membrane from inner side(GO:0032978) protein insertion into mitochondrial membrane from inner side(GO:0032979) mitochondrial respiratory chain complex III assembly(GO:0034551) protein insertion into mitochondrial membrane(GO:0051204) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 | 0.3 | GO:0071962 | mitotic sister chromatid cohesion, centromeric(GO:0071962) |
| 0.1 | 0.4 | GO:0045687 | positive regulation of gliogenesis(GO:0014015) positive regulation of glial cell differentiation(GO:0045687) positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.1 | 1.4 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.1 | 0.2 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.1 | 2.8 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.1 | 0.5 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 2.0 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.1 | 3.5 | GO:0006405 | RNA export from nucleus(GO:0006405) |
| 0.1 | 0.3 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.1 | 0.8 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.1 | 1.0 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 1.3 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.1 | 0.3 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 0.3 | GO:0002082 | regulation of oxidative phosphorylation(GO:0002082) |
| 0.1 | 0.6 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.6 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.1 | 0.3 | GO:0030329 | prenylated protein catabolic process(GO:0030327) prenylcysteine catabolic process(GO:0030328) prenylcysteine metabolic process(GO:0030329) |
| 0.1 | 1.3 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.1 | 2.8 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.1 | 1.1 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 | 1.4 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.1 | 0.2 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.0 | 0.3 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.7 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.0 | GO:0002902 | B cell apoptotic process(GO:0001783) regulation of B cell apoptotic process(GO:0002902) regulation of lymphocyte apoptotic process(GO:0070228) |
| 0.0 | 0.3 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.3 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.0 | 2.2 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.0 | 0.6 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.3 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.3 | GO:0006537 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.0 | 0.3 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.0 | 0.3 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.9 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 1.3 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 1.0 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.5 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.9 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.4 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.2 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 3.5 | GO:0030837 | negative regulation of actin filament polymerization(GO:0030837) negative regulation of protein polymerization(GO:0032272) |
| 0.0 | 2.2 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 0.4 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 1.1 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 1.2 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 2.2 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.4 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.8 | GO:0071174 | mitotic spindle assembly checkpoint(GO:0007094) spindle checkpoint(GO:0031577) negative regulation of mitotic metaphase/anaphase transition(GO:0045841) spindle assembly checkpoint(GO:0071173) mitotic spindle checkpoint(GO:0071174) |
| 0.0 | 0.7 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.8 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.5 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 2.5 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.0 | 1.9 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.2 | GO:0031179 | peptide modification(GO:0031179) |
| 0.0 | 1.6 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.5 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.9 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.1 | GO:0038158 | granulocyte colony-stimulating factor signaling pathway(GO:0038158) |
| 0.0 | 0.4 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 0.1 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.1 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.0 | 0.9 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.1 | GO:0071047 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) nuclear polyadenylation-dependent mRNA catabolic process(GO:0071042) polyadenylation-dependent mRNA catabolic process(GO:0071047) |
| 0.0 | 0.9 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.6 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.5 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.0 | 0.4 | GO:0051084 | 'de novo' protein folding(GO:0006458) 'de novo' posttranslational protein folding(GO:0051084) chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.5 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.1 | GO:0001961 | positive regulation of cytokine-mediated signaling pathway(GO:0001961) positive regulation of response to cytokine stimulus(GO:0060760) |
| 0.0 | 0.2 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 1.4 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.2 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.1 | GO:0003403 | optic vesicle formation(GO:0003403) |
| 0.0 | 0.1 | GO:0033499 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.0 | 0.5 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 1.9 | GO:0070121 | Kupffer's vesicle development(GO:0070121) |
| 0.0 | 0.2 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.7 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.6 | GO:0043631 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.0 | 2.2 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.4 | GO:0070646 | protein modification by small protein removal(GO:0070646) |
| 0.0 | 0.6 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.0 | 1.4 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 0.2 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.0 | 1.4 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.3 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.0 | 0.3 | GO:0005980 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.1 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.5 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 1.3 | GO:2001020 | regulation of response to DNA damage stimulus(GO:2001020) |
| 0.0 | 0.4 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 2.0 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.2 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 0.3 | GO:0071594 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.0 | 0.3 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.4 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) |
| 0.0 | 0.2 | GO:0006032 | chitin catabolic process(GO:0006032) |
| 0.0 | 0.2 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 1.0 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 0.1 | GO:0010390 | histone monoubiquitination(GO:0010390) |
| 0.0 | 0.3 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.2 | GO:0070897 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) DNA-templated transcriptional preinitiation complex assembly(GO:0070897) |
| 0.0 | 0.3 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.1 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.0 | 0.1 | GO:0019682 | glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 0.0 | 2.6 | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process(GO:0043161) |
| 0.0 | 0.3 | GO:0038034 | signal transduction in absence of ligand(GO:0038034) extrinsic apoptotic signaling pathway in absence of ligand(GO:0097192) |
| 0.0 | 3.7 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
| 0.0 | 0.8 | GO:0030100 | regulation of endocytosis(GO:0030100) |
| 0.0 | 0.9 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.3 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 0.0 | GO:0032387 | negative regulation of intracellular transport(GO:0032387) |
| 0.0 | 0.7 | GO:0072583 | clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.3 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.1 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.2 | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.1 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.3 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.4 | GO:0006612 | protein targeting to membrane(GO:0006612) |
| 0.0 | 0.3 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.0 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.7 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.5 | 2.7 | GO:0034657 | GID complex(GO:0034657) |
| 0.4 | 1.2 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.3 | 0.8 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.3 | 2.2 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.3 | 3.7 | GO:0002102 | podosome(GO:0002102) |
| 0.2 | 1.0 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.2 | 1.4 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.2 | 3.4 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.2 | 1.0 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.2 | 1.3 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.2 | 0.6 | GO:1990498 | mitotic spindle microtubule(GO:1990498) |
| 0.2 | 2.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 1.9 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.1 | 0.4 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.5 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 2.5 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 1.0 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 1.7 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 1.9 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 2.0 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 2.6 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 0.7 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 0.5 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.1 | 1.0 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 1.5 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.3 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.1 | 2.0 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 1.2 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 3.5 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 0.6 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 0.6 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 0.5 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 1.3 | GO:0031082 | BLOC complex(GO:0031082) |
| 0.0 | 0.7 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.3 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 1.0 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 1.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.5 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.4 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 2.3 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.3 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.4 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 1.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.8 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 2.2 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.6 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.5 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 1.9 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.2 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.3 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.4 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 0.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 1.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.2 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.0 | 1.3 | GO:0016529 | sarcoplasm(GO:0016528) sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.4 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.6 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.3 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 3.0 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 1.2 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.2 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 1.1 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 1.0 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.5 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.0 | 0.1 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 2.5 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.1 | GO:0031261 | DNA replication preinitiation complex(GO:0031261) |
| 0.0 | 1.9 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 1.6 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 1.0 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.9 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 1.4 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.5 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.6 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 2.0 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 2.2 | GO:0099572 | postsynaptic specialization(GO:0099572) |
| 0.0 | 0.8 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 1.1 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.7 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 1.1 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.5 | GO:0048475 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
| 0.0 | 0.2 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.1 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 1.3 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.1 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 1.0 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 0.3 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.5 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.6 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.3 | GO:0032589 | neuron projection membrane(GO:0032589) dendrite membrane(GO:0032590) |
| 0.0 | 0.8 | GO:0031201 | SNARE complex(GO:0031201) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.5 | 1.6 | GO:0061599 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.5 | 1.5 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.5 | 2.0 | GO:0070404 | 6,7-dihydropteridine reductase activity(GO:0004155) NADH binding(GO:0070404) |
| 0.5 | 3.4 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.5 | 1.4 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.5 | 2.3 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.4 | 1.2 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.4 | 3.3 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.4 | 1.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.4 | 1.4 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.3 | 3.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.3 | 2.8 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.3 | 1.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.3 | 2.3 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.2 | 0.7 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.2 | 1.4 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.2 | 2.5 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.2 | 2.2 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.2 | 1.5 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.2 | 0.8 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.2 | 2.0 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.2 | 0.9 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) RNA polymerase I core binding(GO:0001042) |
| 0.2 | 2.3 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.2 | 1.4 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.2 | 1.0 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.2 | 1.9 | GO:0043495 | protein anchor(GO:0043495) |
| 0.2 | 0.8 | GO:0019778 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.2 | 2.5 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.2 | 0.6 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 1.9 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 1.9 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 2.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 0.6 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.1 | 1.2 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.1 | 1.1 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 2.1 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.1 | 0.6 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 3.6 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 0.5 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.1 | 0.6 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.1 | 2.1 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 0.7 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 1.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 1.6 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 0.7 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 0.3 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.1 | 0.3 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.1 | 0.4 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.1 | 0.6 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 0.4 | GO:0004307 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.1 | 0.3 | GO:0030882 | lipid antigen binding(GO:0030882) |
| 0.1 | 1.9 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 1.2 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.1 | 7.5 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 0.5 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 1.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 1.1 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.1 | 0.3 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.1 | 1.4 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.4 | GO:0004997 | thyrotropin-releasing hormone receptor activity(GO:0004997) |
| 0.1 | 1.0 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 0.3 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.1 | 0.3 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.1 | 0.9 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 2.7 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 0.2 | GO:0070905 | glycine hydroxymethyltransferase activity(GO:0004372) serine binding(GO:0070905) |
| 0.1 | 1.1 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 2.6 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 3.7 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 0.5 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 0.3 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 1.0 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.3 | GO:0008469 | histone-arginine N-methyltransferase activity(GO:0008469) |
| 0.1 | 0.3 | GO:0001735 | prenylcysteine oxidase activity(GO:0001735) |
| 0.1 | 1.2 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 0.5 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.1 | 0.3 | GO:0008184 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 0.2 | GO:0008117 | sphinganine-1-phosphate aldolase activity(GO:0008117) |
| 0.1 | 0.6 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 1.5 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 1.0 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.2 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.7 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.4 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.2 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 1.4 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 2.1 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 1.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 1.4 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.5 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.1 | GO:0003978 | UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.0 | 0.3 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 1.0 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.2 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.0 | 0.3 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.3 | GO:0008515 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.3 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.5 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 1.4 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.3 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 6.2 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 4.0 | GO:0019783 | ubiquitin-like protein-specific protease activity(GO:0019783) |
| 0.0 | 1.5 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 2.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.8 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.2 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.0 | 0.9 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 0.2 | GO:0001047 | core promoter sequence-specific DNA binding(GO:0001046) core promoter binding(GO:0001047) |
| 0.0 | 0.3 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.2 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.2 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.2 | GO:0000048 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.9 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.9 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 2.3 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.4 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.2 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 2.2 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.4 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 2.2 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.7 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.4 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 1.2 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.2 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.4 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.3 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.4 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.9 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.2 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 1.0 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.6 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 0.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.9 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.6 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 5.8 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 1.1 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 0.1 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 1.1 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.2 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.6 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 1.2 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.5 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.7 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 1.1 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.3 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 3.8 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 1.5 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 1.3 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 5.7 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 1.4 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.1 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.3 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 1.2 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.1 | 1.8 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 2.0 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 0.3 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.1 | 0.6 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.1 | 1.8 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 1.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 0.2 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.1 | 0.7 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 0.8 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 1.0 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 3.8 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 3.1 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 1.8 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 2.2 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 1.2 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.4 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.5 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.5 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.3 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.4 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.8 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 1.0 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.2 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.9 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.6 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.3 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.2 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.2 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.2 | PID FOXO PATHWAY | FoxO family signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.7 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.1 | 1.5 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 1.1 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 1.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.4 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 1.8 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 1.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 1.1 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 1.6 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 4.0 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.1 | 3.3 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 3.1 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 0.8 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 0.9 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 1.5 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 0.9 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 2.2 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 2.3 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 1.0 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 1.4 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.9 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.8 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.9 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 7.7 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.3 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.3 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 1.3 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 1.3 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.6 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.3 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.0 | 0.5 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.1 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.2 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 0.5 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.0 | 0.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.3 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.5 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.2 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.0 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 0.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.8 | REACTOME TRAF6 MEDIATED INDUCTION OF NFKB AND MAP KINASES UPON TLR7 8 OR 9 ACTIVATION | Genes involved in TRAF6 mediated induction of NFkB and MAP kinases upon TLR7/8 or 9 activation |
| 0.0 | 0.3 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.2 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.3 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.6 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |