Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
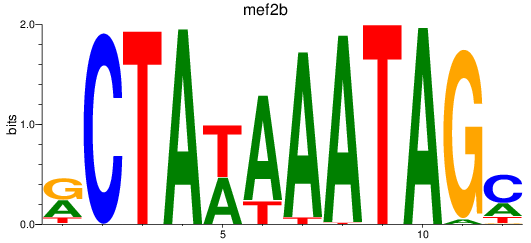
Results for mef2b
Z-value: 1.32
Transcription factors associated with mef2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mef2b
|
ENSDARG00000093170 | myocyte enhancer factor 2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mef2b | dr11_v1_chr22_+_18188045_18188045 | 0.46 | 2.1e-01 | Click! |
Activity profile of mef2b motif
Sorted Z-values of mef2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_61205711 | 5.10 |
ENSDART00000055062
|
pvalb1
|
parvalbumin 1 |
| chr3_-_32818607 | 4.18 |
ENSDART00000075465
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr25_+_29160102 | 3.38 |
ENSDART00000162854
|
pkmb
|
pyruvate kinase M1/2b |
| chr11_+_11201096 | 2.85 |
ENSDART00000171916
ENSDART00000171521 ENSDART00000087105 ENSDART00000159603 |
myom2a
|
myomesin 2a |
| chr9_-_33107237 | 2.70 |
ENSDART00000013918
|
casq2
|
calsequestrin 2 |
| chr5_-_72125551 | 2.65 |
ENSDART00000149412
|
smyd1a
|
SET and MYND domain containing 1a |
| chr16_+_29663809 | 2.41 |
ENSDART00000191336
|
tmod4
|
tropomodulin 4 (muscle) |
| chr6_-_42003780 | 2.39 |
ENSDART00000032527
|
cav3
|
caveolin 3 |
| chr5_+_51597677 | 2.31 |
ENSDART00000048210
ENSDART00000184797 |
ckmt2b
|
creatine kinase, mitochondrial 2b (sarcomeric) |
| chr23_+_20110086 | 2.12 |
ENSDART00000054664
|
tnnc1b
|
troponin C type 1b (slow) |
| chr10_-_8032885 | 2.04 |
ENSDART00000188619
|
atp6v0a2a
|
ATPase H+ transporting V0 subunit a2a |
| chr20_-_26001288 | 1.90 |
ENSDART00000136518
ENSDART00000063177 |
capn3b
|
calpain 3b |
| chr8_+_22930627 | 1.85 |
ENSDART00000187860
|
sypa
|
synaptophysin a |
| chr13_+_9432501 | 1.85 |
ENSDART00000058064
|
zgc:123321
|
zgc:123321 |
| chr10_-_22803740 | 1.84 |
ENSDART00000079469
ENSDART00000187968 ENSDART00000122543 |
pcolcea
|
procollagen C-endopeptidase enhancer a |
| chr6_-_14139503 | 1.81 |
ENSDART00000089577
|
cacnb4b
|
calcium channel, voltage-dependent, beta 4b subunit |
| chr10_-_8033468 | 1.76 |
ENSDART00000140476
|
atp6v0a2a
|
ATPase H+ transporting V0 subunit a2a |
| chr17_-_10025234 | 1.76 |
ENSDART00000008355
|
cfl2
|
cofilin 2 (muscle) |
| chr1_+_45080897 | 1.59 |
ENSDART00000129819
|
si:ch211-151p13.8
|
si:ch211-151p13.8 |
| chr23_+_22658700 | 1.53 |
ENSDART00000192248
|
eno1a
|
enolase 1a, (alpha) |
| chr9_+_31795343 | 1.52 |
ENSDART00000139584
|
itgbl1
|
integrin, beta-like 1 |
| chr17_-_12336987 | 1.48 |
ENSDART00000172001
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr5_+_67390645 | 1.41 |
ENSDART00000014822
|
ebf2
|
early B cell factor 2 |
| chr10_+_37145007 | 1.38 |
ENSDART00000131777
|
cuedc1a
|
CUE domain containing 1a |
| chr20_-_26042070 | 1.37 |
ENSDART00000140255
|
si:dkey-12h9.6
|
si:dkey-12h9.6 |
| chr5_-_36948586 | 1.33 |
ENSDART00000193606
|
h3f3c
|
H3 histone, family 3C |
| chr10_-_24371312 | 1.33 |
ENSDART00000149362
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr20_+_15552657 | 1.29 |
ENSDART00000063912
|
jun
|
Jun proto-oncogene, AP-1 transcription factor subunit |
| chr1_+_10683843 | 1.26 |
ENSDART00000054879
|
zgc:103678
|
zgc:103678 |
| chr8_+_29962635 | 1.24 |
ENSDART00000007640
|
ptch1
|
patched 1 |
| chr12_+_6002715 | 1.24 |
ENSDART00000114961
|
si:ch211-131k2.3
|
si:ch211-131k2.3 |
| chr4_-_23908802 | 1.19 |
ENSDART00000138873
|
celf2
|
cugbp, Elav-like family member 2 |
| chr19_-_41069573 | 1.19 |
ENSDART00000111982
ENSDART00000193142 |
sgce
|
sarcoglycan, epsilon |
| chr1_-_25936677 | 1.17 |
ENSDART00000146488
ENSDART00000136321 |
myoz2b
|
myozenin 2b |
| chr5_-_31875645 | 1.16 |
ENSDART00000098160
|
tmem119b
|
transmembrane protein 119b |
| chr5_+_67390115 | 1.15 |
ENSDART00000193255
|
ebf2
|
early B cell factor 2 |
| chr7_+_14291323 | 1.15 |
ENSDART00000053521
|
rhcga
|
Rh family, C glycoprotein a |
| chr14_+_24215046 | 1.15 |
ENSDART00000079215
|
stc2a
|
stanniocalcin 2a |
| chr25_+_3677650 | 1.14 |
ENSDART00000154348
|
prnprs3
|
prion protein, related sequence 3 |
| chr10_-_22845485 | 1.12 |
ENSDART00000079454
|
vamp2
|
vesicle-associated membrane protein 2 |
| chr5_+_64368770 | 1.09 |
ENSDART00000162246
|
plpp7
|
phospholipid phosphatase 7 (inactive) |
| chr21_+_19070921 | 1.06 |
ENSDART00000029874
|
nkx6.1
|
NK6 homeobox 1 |
| chr20_+_34455645 | 1.02 |
ENSDART00000135789
|
mettl11b
|
methyltransferase like 11B |
| chr19_-_38611814 | 1.01 |
ENSDART00000151958
|
col16a1
|
collagen, type XVI, alpha 1 |
| chr24_-_32665283 | 1.00 |
ENSDART00000038364
|
ca2
|
carbonic anhydrase II |
| chr24_-_20444844 | 1.00 |
ENSDART00000048940
|
vill
|
villin-like |
| chr24_+_25471196 | 1.00 |
ENSDART00000066625
|
smpx
|
small muscle protein, X-linked |
| chr11_-_18253111 | 0.99 |
ENSDART00000125984
|
mustn1b
|
musculoskeletal, embryonic nuclear protein 1b |
| chr12_+_8373525 | 0.95 |
ENSDART00000152180
|
arid5b
|
AT-rich interaction domain 5B |
| chr1_+_8601935 | 0.95 |
ENSDART00000152367
|
si:ch211-160d14.6
|
si:ch211-160d14.6 |
| chr19_+_41169996 | 0.93 |
ENSDART00000048438
|
asb4
|
ankyrin repeat and SOCS box containing 4 |
| chr25_-_13381854 | 0.91 |
ENSDART00000164621
ENSDART00000169129 |
ndrg4
|
NDRG family member 4 |
| chr5_-_51819027 | 0.88 |
ENSDART00000164267
|
homer1b
|
homer scaffolding protein 1b |
| chr23_+_23485858 | 0.84 |
ENSDART00000114067
|
agrn
|
agrin |
| chr7_-_27686021 | 0.82 |
ENSDART00000079112
ENSDART00000100989 |
calca
|
calcitonin/calcitonin-related polypeptide, alpha |
| chr10_-_4980150 | 0.81 |
ENSDART00000093228
|
mat2al
|
methionine adenosyltransferase II, alpha-like |
| chr21_-_12272543 | 0.78 |
ENSDART00000081510
ENSDART00000151297 |
celf4
|
CUGBP, Elav-like family member 4 |
| chr12_-_34035364 | 0.76 |
ENSDART00000087065
|
timp2a
|
TIMP metallopeptidase inhibitor 2a |
| chr6_-_10780698 | 0.75 |
ENSDART00000151714
|
gpr155b
|
G protein-coupled receptor 155b |
| chr14_+_35901249 | 0.74 |
ENSDART00000105604
|
zgc:77938
|
zgc:77938 |
| chr6_-_40722200 | 0.73 |
ENSDART00000035101
|
kbtbd12
|
kelch repeat and BTB (POZ) domain containing 12 |
| chr2_-_33993533 | 0.73 |
ENSDART00000140910
ENSDART00000077304 |
ptch2
|
patched 2 |
| chr6_-_40722480 | 0.72 |
ENSDART00000188187
|
kbtbd12
|
kelch repeat and BTB (POZ) domain containing 12 |
| chr14_-_32405387 | 0.69 |
ENSDART00000184647
|
fgf13a
|
fibroblast growth factor 13a |
| chr24_+_8736497 | 0.66 |
ENSDART00000181904
|
tmem14ca
|
transmembrane protein 14Ca |
| chr23_-_39849155 | 0.65 |
ENSDART00000115330
|
ppp1r14c
|
protein phosphatase 1, regulatory (inhibitor) subunit 14C |
| chr7_-_7420301 | 0.65 |
ENSDART00000102620
|
six7
|
SIX homeobox 7 |
| chr12_+_34119439 | 0.63 |
ENSDART00000032821
|
cyth1b
|
cytohesin 1b |
| chr1_-_17569793 | 0.63 |
ENSDART00000125125
|
acsl1a
|
acyl-CoA synthetase long chain family member 1a |
| chr8_-_25002182 | 0.62 |
ENSDART00000078792
|
ahcyl1
|
adenosylhomocysteinase-like 1 |
| chr11_-_39202915 | 0.62 |
ENSDART00000105133
|
wnt4a
|
wingless-type MMTV integration site family, member 4a |
| chr12_+_30563550 | 0.60 |
ENSDART00000126064
|
si:ch211-28p3.4
|
si:ch211-28p3.4 |
| chr1_-_44638058 | 0.59 |
ENSDART00000081835
|
slc43a1b
|
solute carrier family 43 (amino acid system L transporter), member 1b |
| chr13_-_50139916 | 0.59 |
ENSDART00000099475
|
nid1a
|
nidogen 1a |
| chr23_+_44614056 | 0.55 |
ENSDART00000188379
|
eno3
|
enolase 3, (beta, muscle) |
| chr1_-_411331 | 0.54 |
ENSDART00000092524
|
rasa3
|
RAS p21 protein activator 3 |
| chr22_+_9772754 | 0.54 |
ENSDART00000130194
|
BX664625.2
|
|
| chr7_-_23745984 | 0.54 |
ENSDART00000048050
|
ITGB1BP2
|
zgc:92429 |
| chr13_+_42011287 | 0.54 |
ENSDART00000131147
|
cyp1b1
|
cytochrome P450, family 1, subfamily B, polypeptide 1 |
| chr3_+_23092762 | 0.53 |
ENSDART00000142884
ENSDART00000024136 |
gngt2a
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2a |
| chr17_-_13026634 | 0.53 |
ENSDART00000113713
|
fam177a1
|
family with sequence similarity 177, member A1 |
| chr8_+_44420108 | 0.53 |
ENSDART00000075381
|
CU571323.1
|
|
| chr2_+_19777146 | 0.52 |
ENSDART00000038648
|
ptbp2b
|
polypyrimidine tract binding protein 2b |
| chr14_-_24332786 | 0.52 |
ENSDART00000173164
|
fam13b
|
family with sequence similarity 13, member B |
| chr14_+_21107032 | 0.51 |
ENSDART00000138319
ENSDART00000139103 ENSDART00000184735 |
aldob
|
aldolase b, fructose-bisphosphate |
| chr6_-_35472923 | 0.49 |
ENSDART00000185907
|
rgs8
|
regulator of G protein signaling 8 |
| chr2_+_1202347 | 0.48 |
ENSDART00000075837
|
CABZ01084566.1
|
|
| chr10_+_29259882 | 0.48 |
ENSDART00000180606
|
sytl2a
|
synaptotagmin-like 2a |
| chr5_+_64842730 | 0.46 |
ENSDART00000144732
|
lrrc8ab
|
leucine rich repeat containing 8 VRAC subunit Ab |
| chr25_-_21816269 | 0.46 |
ENSDART00000152014
|
zgc:158222
|
zgc:158222 |
| chr21_+_11685009 | 0.45 |
ENSDART00000014668
|
pcsk1
|
proprotein convertase subtilisin/kexin type 1 |
| chr22_+_26443235 | 0.44 |
ENSDART00000044085
|
zgc:92480
|
zgc:92480 |
| chr1_-_25177086 | 0.44 |
ENSDART00000144711
ENSDART00000177225 |
tmem154
|
transmembrane protein 154 |
| chr17_+_6956696 | 0.44 |
ENSDART00000171368
|
zgc:172341
|
zgc:172341 |
| chr12_+_42400277 | 0.44 |
ENSDART00000166590
|
si:ch211-221j21.3
|
si:ch211-221j21.3 |
| chr16_+_16969060 | 0.43 |
ENSDART00000182819
ENSDART00000191876 |
si:ch211-120k19.1
rpl18
|
si:ch211-120k19.1 ribosomal protein L18 |
| chr5_-_10946232 | 0.43 |
ENSDART00000163139
ENSDART00000031265 |
rtn4r
|
reticulon 4 receptor |
| chr21_+_11684830 | 0.42 |
ENSDART00000147473
|
pcsk1
|
proprotein convertase subtilisin/kexin type 1 |
| chr1_-_17570013 | 0.42 |
ENSDART00000146946
|
acsl1a
|
acyl-CoA synthetase long chain family member 1a |
| chr16_+_16968682 | 0.42 |
ENSDART00000111074
|
si:ch211-120k19.1
|
si:ch211-120k19.1 |
| chr12_-_36740781 | 0.41 |
ENSDART00000105484
|
si:ch211-216b21.2
|
si:ch211-216b21.2 |
| chr21_+_12010505 | 0.40 |
ENSDART00000123522
|
aqp7
|
aquaporin 7 |
| chr15_-_14552101 | 0.40 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr10_+_29260096 | 0.40 |
ENSDART00000088973
|
sytl2a
|
synaptotagmin-like 2a |
| chr9_-_48281941 | 0.40 |
ENSDART00000099787
|
klhl41a
|
kelch-like family member 41a |
| chr8_+_17987215 | 0.40 |
ENSDART00000113605
|
lrriq3
|
leucine-rich repeats and IQ motif containing 3 |
| chr9_-_48937240 | 0.39 |
ENSDART00000075627
|
cers6
|
ceramide synthase 6 |
| chr7_-_52334840 | 0.38 |
ENSDART00000174173
|
CR938716.1
|
|
| chr24_-_4450238 | 0.38 |
ENSDART00000066835
|
fzd8a
|
frizzled class receptor 8a |
| chr8_-_50259448 | 0.37 |
ENSDART00000146056
|
nkx3-1
|
NK3 homeobox 1 |
| chr21_-_25669820 | 0.36 |
ENSDART00000148236
|
tmem179b
|
transmembrane protein 179B |
| chr12_+_41697664 | 0.36 |
ENSDART00000162302
|
bnip3
|
BCL2 interacting protein 3 |
| chr24_-_12745222 | 0.36 |
ENSDART00000151836
|
si:ch211-196f5.9
|
si:ch211-196f5.9 |
| chr17_+_6957042 | 0.34 |
ENSDART00000103839
|
zgc:172341
|
zgc:172341 |
| chr2_+_37975026 | 0.34 |
ENSDART00000034802
|
si:rp71-1g18.13
|
si:rp71-1g18.13 |
| chr21_+_25669934 | 0.32 |
ENSDART00000101203
|
sumf2
|
sulfatase modifying factor 2 |
| chr1_-_21483832 | 0.32 |
ENSDART00000102790
|
glrba
|
glycine receptor, beta a |
| chr10_+_29698467 | 0.31 |
ENSDART00000163402
|
dlg2
|
discs, large homolog 2 (Drosophila) |
| chr19_-_31035155 | 0.30 |
ENSDART00000161882
|
bzw2
|
basic leucine zipper and W2 domains 2 |
| chr19_+_24394560 | 0.28 |
ENSDART00000142506
|
si:dkey-81h8.1
|
si:dkey-81h8.1 |
| chr12_-_26430507 | 0.27 |
ENSDART00000153214
|
synpo2lb
|
synaptopodin 2-like b |
| chr3_-_52674089 | 0.27 |
ENSDART00000154260
ENSDART00000125455 |
si:dkey-210j14.4
|
si:dkey-210j14.4 |
| chr1_-_6494384 | 0.26 |
ENSDART00000109356
|
klf7a
|
Kruppel-like factor 7a |
| chr23_+_44497701 | 0.25 |
ENSDART00000149903
|
si:ch73-375g18.1
|
si:ch73-375g18.1 |
| chr16_-_17200120 | 0.24 |
ENSDART00000147739
|
gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr25_-_29363934 | 0.24 |
ENSDART00000166889
|
nptna
|
neuroplastin a |
| chr18_+_7264961 | 0.23 |
ENSDART00000188461
|
CABZ01015105.1
|
|
| chr4_-_5863906 | 0.22 |
ENSDART00000169424
|
slc5a8
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 8 |
| chr4_-_9637398 | 0.22 |
ENSDART00000150253
|
dclre1c
|
DNA cross-link repair 1C, PSO2 homolog (S. cerevisiae) |
| chr19_-_31035325 | 0.22 |
ENSDART00000147504
|
bzw2
|
basic leucine zipper and W2 domains 2 |
| chr25_-_29988352 | 0.21 |
ENSDART00000067059
|
fam19a5b
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5b |
| chr7_+_33457148 | 0.21 |
ENSDART00000133562
ENSDART00000074587 |
paqr5b
|
progestin and adipoQ receptor family member Vb |
| chr9_-_28399071 | 0.20 |
ENSDART00000104317
ENSDART00000064343 |
klf7b
|
Kruppel-like factor 7b |
| chr10_-_17170086 | 0.20 |
ENSDART00000020122
|
ywhah
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide |
| chr25_-_27722614 | 0.20 |
ENSDART00000190154
|
zgc:153935
|
zgc:153935 |
| chr20_-_26383368 | 0.19 |
ENSDART00000024518
ENSDART00000087844 |
esr1
|
estrogen receptor 1 |
| chr6_-_46403475 | 0.19 |
ENSDART00000154148
|
camk1a
|
calcium/calmodulin-dependent protein kinase Ia |
| chr21_-_27185915 | 0.18 |
ENSDART00000135052
|
slc8a4a
|
solute carrier family 8 (sodium/calcium exchanger), member 4a |
| chr4_+_5180650 | 0.17 |
ENSDART00000067390
|
fgf6b
|
fibroblast growth factor 6b |
| chr1_-_45662774 | 0.17 |
ENSDART00000042158
|
serhl
|
serine hydrolase-like |
| chr23_+_6077503 | 0.16 |
ENSDART00000081714
ENSDART00000139834 |
mybpha
|
myosin binding protein Ha |
| chr15_-_36347858 | 0.15 |
ENSDART00000155274
ENSDART00000157936 |
si:dkey-23k10.2
|
si:dkey-23k10.2 |
| chr23_+_16889352 | 0.14 |
ENSDART00000145275
|
si:dkey-147f3.8
|
si:dkey-147f3.8 |
| chr9_+_29040425 | 0.13 |
ENSDART00000150201
|
MRAS
|
si:ch73-116o1.2 |
| chr23_-_25050329 | 0.13 |
ENSDART00000140216
|
avpr2aa
|
arginine vasopressin receptor 2a, duplicate a |
| chr18_+_40354998 | 0.13 |
ENSDART00000098791
ENSDART00000049171 |
sema6dl
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D, like |
| chr19_-_32518556 | 0.12 |
ENSDART00000103410
|
zbtb8b
|
zinc finger and BTB domain containing 8B |
| chr25_-_27722309 | 0.12 |
ENSDART00000148121
|
zgc:153935
|
zgc:153935 |
| chr17_-_15600455 | 0.11 |
ENSDART00000110272
ENSDART00000156911 |
si:ch211-266g18.9
|
si:ch211-266g18.9 |
| chr21_+_43559123 | 0.11 |
ENSDART00000151212
|
gpr185a
|
G protein-coupled receptor 185 a |
| chr17_-_50234004 | 0.11 |
ENSDART00000058706
|
fosaa
|
v-fos FBJ murine osteosarcoma viral oncogene homolog Aa |
| chr4_+_77060861 | 0.10 |
ENSDART00000174271
ENSDART00000174393 ENSDART00000150450 |
si:dkey-240n22.8
|
si:dkey-240n22.8 |
| chr7_+_21859337 | 0.10 |
ENSDART00000159626
|
si:dkey-85k7.7
|
si:dkey-85k7.7 |
| chr17_-_14671098 | 0.10 |
ENSDART00000037371
|
ppp1r13ba
|
protein phosphatase 1, regulatory subunit 13Ba |
| chr22_-_26852516 | 0.09 |
ENSDART00000005829
|
gde1
|
glycerophosphodiester phosphodiesterase 1 |
| chr19_+_44039849 | 0.09 |
ENSDART00000086040
|
lrrc14b
|
leucine rich repeat containing 14B |
| chr17_-_26867725 | 0.09 |
ENSDART00000153590
|
si:dkey-221l4.10
|
si:dkey-221l4.10 |
| chr22_+_38310957 | 0.08 |
ENSDART00000040550
|
traf5
|
Tnf receptor-associated factor 5 |
| chr24_+_26088143 | 0.08 |
ENSDART00000171129
|
CR352209.1
|
|
| chr22_-_4769140 | 0.07 |
ENSDART00000165235
|
calr3a
|
calreticulin 3a |
| chr6_+_36942966 | 0.07 |
ENSDART00000028895
|
negr1
|
neuronal growth regulator 1 |
| chr4_-_13509946 | 0.06 |
ENSDART00000134720
|
ifng1-2
|
interferon, gamma 1-2 |
| chr23_-_7826849 | 0.06 |
ENSDART00000157612
|
myt1b
|
myelin transcription factor 1b |
| chr25_-_19486399 | 0.05 |
ENSDART00000155076
ENSDART00000156016 |
zgc:193812
|
zgc:193812 |
| chr17_+_1992495 | 0.05 |
ENSDART00000192937
|
CABZ01062996.1
|
|
| chr23_+_3721042 | 0.05 |
ENSDART00000143323
|
smim29
|
small integral membrane protein 29 |
| chr17_+_45648836 | 0.05 |
ENSDART00000155037
|
zgc:162184
|
zgc:162184 |
| chr5_-_50781623 | 0.03 |
ENSDART00000114950
|
zgc:194908
|
zgc:194908 |
| chr5_+_44346691 | 0.03 |
ENSDART00000034523
|
tars
|
threonyl-tRNA synthetase |
| chr5_+_16580739 | 0.01 |
ENSDART00000135140
|
htr7c
|
5-hydroxytryptamine (serotonin) receptor 7c |
| chr4_+_14343706 | 0.01 |
ENSDART00000142845
|
prl2
|
prolactin 2 |
| chr13_-_37647209 | 0.01 |
ENSDART00000189102
|
si:dkey-188i13.10
|
si:dkey-188i13.10 |
| chr10_+_8688678 | 0.00 |
ENSDART00000147568
|
si:dkey-27b3.4
|
si:dkey-27b3.4 |
| chr9_-_23894392 | 0.00 |
ENSDART00000133417
|
asb18
|
ankyrin repeat and SOCS box containing 18 |
Network of associatons between targets according to the STRING database.
First level regulatory network of mef2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.7 | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling(GO:0010882) |
| 0.6 | 1.9 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.6 | 1.8 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.4 | 1.2 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.3 | 1.0 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.3 | 0.8 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.3 | 1.4 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.3 | 3.8 | GO:0045851 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.3 | 2.4 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.3 | 2.4 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.3 | 2.6 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.3 | 2.1 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.3 | 1.0 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.2 | 1.1 | GO:0021731 | trigeminal motor nucleus development(GO:0021731) |
| 0.2 | 2.0 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.2 | 1.0 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.2 | 2.3 | GO:0046314 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.2 | 2.4 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 1.2 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.1 | 0.4 | GO:0060898 | spinal cord radial glial cell differentiation(GO:0021531) eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.1 | 0.9 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 0.8 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 | 0.5 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.1 | 0.8 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 0.7 | GO:0046292 | formaldehyde metabolic process(GO:0046292) formaldehyde catabolic process(GO:0046294) |
| 0.1 | 1.0 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.1 | 1.3 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.1 | 1.2 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 | 1.1 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 4.1 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.1 | 1.1 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.1 | 1.0 | GO:0015858 | nucleoside transport(GO:0015858) nucleoside transmembrane transport(GO:1901642) |
| 0.1 | 0.5 | GO:0043363 | nucleate erythrocyte differentiation(GO:0043363) |
| 0.1 | 0.9 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 2.0 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.7 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 1.5 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 1.8 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 1.8 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 1.1 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.5 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.6 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 0.4 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.0 | 1.0 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.8 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.2 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.0 | 0.7 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.2 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) telomere maintenance in response to DNA damage(GO:0043247) |
| 0.0 | 1.3 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.3 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 0.4 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.6 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.2 | GO:0042044 | fluid transport(GO:0042044) |
| 0.0 | 0.6 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.3 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.1 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.4 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 1.4 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.7 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.4 | 3.8 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.3 | 6.5 | GO:0031430 | M band(GO:0031430) |
| 0.3 | 2.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.2 | 1.5 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.2 | 1.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 4.5 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.1 | 1.4 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 2.4 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.4 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 1.5 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.5 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 1.1 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.5 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.2 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 1.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 1.9 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.3 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 1.4 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.7 | GO:0030426 | growth cone(GO:0030426) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.4 | GO:0071253 | connexin binding(GO:0071253) |
| 0.6 | 3.4 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.5 | 2.0 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.3 | 1.0 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.3 | 0.8 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.3 | 2.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.3 | 3.8 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.2 | 0.6 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.2 | 0.8 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.2 | 1.3 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.2 | 2.3 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.2 | 0.9 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 0.7 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) S-(hydroxymethyl)glutathione dehydrogenase activity(GO:0051903) |
| 0.1 | 1.0 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.1 | 1.2 | GO:0031433 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.1 | 4.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 2.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 1.1 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 0.5 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 1.2 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 2.1 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 0.7 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.4 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.1 | 0.5 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 1.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 1.2 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.1 | 1.8 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.2 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 0.8 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.2 | GO:0004377 | GDP-Man:Man3GlcNAc2-PP-Dol alpha-1,2-mannosyltransferase activity(GO:0004377) |
| 0.0 | 0.4 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 2.6 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.3 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.9 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.2 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.0 | 0.2 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.4 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 1.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.4 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.5 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 1.3 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.4 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 1.0 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.6 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 4.6 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 13.5 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.1 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 1.2 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 2.4 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 2.0 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 1.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.1 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.7 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.1 | 0.4 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 1.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 0.9 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.1 | 1.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 0.5 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 0.8 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 2.8 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 1.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |