Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
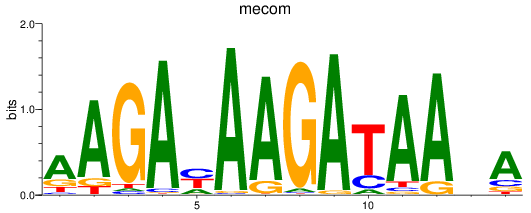
Results for mecom
Z-value: 1.86
Transcription factors associated with mecom
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mecom
|
ENSDARG00000060808 | MDS1 and EVI1 complex locus |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mecom | dr11_v1_chr15_-_35406564_35406564 | 0.95 | 1.2e-04 | Click! |
Activity profile of mecom motif
Sorted Z-values of mecom motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_61203203 | 5.40 |
ENSDART00000171787
|
pvalb1
|
parvalbumin 1 |
| chr3_-_32817274 | 4.65 |
ENSDART00000142582
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr17_-_125091 | 4.22 |
ENSDART00000158825
|
actc1b
|
actin, alpha, cardiac muscle 1b |
| chr16_-_41990421 | 3.66 |
ENSDART00000055921
|
pycard
|
PYD and CARD domain containing |
| chr21_+_6556635 | 3.56 |
ENSDART00000139598
|
col5a1
|
procollagen, type V, alpha 1 |
| chr7_-_8374950 | 3.54 |
ENSDART00000057101
|
aep1
|
aerolysin-like protein |
| chr10_-_22854758 | 3.52 |
ENSDART00000079449
|
and3
|
actinodin3 |
| chr15_-_23647078 | 3.24 |
ENSDART00000059366
|
ckmb
|
creatine kinase, muscle b |
| chr5_-_20195350 | 3.09 |
ENSDART00000139675
|
dao.1
|
D-amino-acid oxidase, tandem duplicate 1 |
| chr13_+_13681681 | 2.89 |
ENSDART00000057825
|
cfd
|
complement factor D (adipsin) |
| chr18_-_6633984 | 2.81 |
ENSDART00000185241
|
tnni1c
|
troponin I, skeletal, slow c |
| chr21_+_7582036 | 2.75 |
ENSDART00000135485
ENSDART00000027268 |
otpa
|
orthopedia homeobox a |
| chr7_-_24364536 | 2.73 |
ENSDART00000064789
|
txn
|
thioredoxin |
| chr13_-_33822550 | 2.72 |
ENSDART00000143703
|
flrt3
|
fibronectin leucine rich transmembrane 3 |
| chr22_-_17611742 | 2.61 |
ENSDART00000144031
|
gpx4a
|
glutathione peroxidase 4a |
| chr15_+_19838458 | 2.51 |
ENSDART00000101204
|
alcamb
|
activated leukocyte cell adhesion molecule b |
| chr15_+_22311803 | 2.39 |
ENSDART00000150182
|
hepacama
|
hepatic and glial cell adhesion molecule a |
| chr14_+_35691889 | 2.36 |
ENSDART00000074685
|
glrbb
|
glycine receptor, beta b |
| chr23_+_20110086 | 2.33 |
ENSDART00000054664
|
tnnc1b
|
troponin C type 1b (slow) |
| chr7_+_38750871 | 2.29 |
ENSDART00000114238
ENSDART00000052325 ENSDART00000137001 |
f2
|
coagulation factor II (thrombin) |
| chr10_-_31782616 | 2.28 |
ENSDART00000128839
|
fez1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr23_+_44745317 | 2.24 |
ENSDART00000165654
|
atp1b2a
|
ATPase Na+/K+ transporting subunit beta 2a |
| chr25_+_18564266 | 2.03 |
ENSDART00000172338
|
cav1
|
caveolin 1 |
| chr5_+_49744713 | 1.98 |
ENSDART00000133384
|
nr2f1a
|
nuclear receptor subfamily 2, group F, member 1a |
| chr18_+_16246806 | 1.96 |
ENSDART00000142584
|
alx1
|
ALX homeobox 1 |
| chr3_-_18575868 | 1.86 |
ENSDART00000122968
|
aqp8b
|
aquaporin 8b |
| chr20_+_26538137 | 1.84 |
ENSDART00000045397
|
stx11b.1
|
syntaxin 11b, tandem duplicate 1 |
| chr15_+_33991928 | 1.84 |
ENSDART00000170177
|
vwde
|
von Willebrand factor D and EGF domains |
| chr24_+_25069609 | 1.82 |
ENSDART00000115165
|
amer2
|
APC membrane recruitment protein 2 |
| chr15_+_19646902 | 1.81 |
ENSDART00000179822
|
lim2.3
|
lens intrinsic membrane protein 2.3 |
| chr14_+_33722950 | 1.80 |
ENSDART00000075312
|
apln
|
apelin |
| chr2_-_20923864 | 1.73 |
ENSDART00000006870
|
ptgs2a
|
prostaglandin-endoperoxide synthase 2a |
| chr25_+_18563476 | 1.72 |
ENSDART00000170841
|
cav1
|
caveolin 1 |
| chr9_+_48455909 | 1.67 |
ENSDART00000167243
|
lrp2a
|
low density lipoprotein receptor-related protein 2a |
| chr3_+_23743139 | 1.67 |
ENSDART00000187409
|
hoxb3a
|
homeobox B3a |
| chr13_+_10232695 | 1.65 |
ENSDART00000080805
|
six2a
|
SIX homeobox 2a |
| chr18_-_6634424 | 1.62 |
ENSDART00000062423
ENSDART00000179955 |
tnni1c
|
troponin I, skeletal, slow c |
| chr8_-_14179798 | 1.59 |
ENSDART00000040645
ENSDART00000146749 |
rhoaa
|
ras homolog gene family, member Aa |
| chr10_-_28761454 | 1.57 |
ENSDART00000129400
|
alcama
|
activated leukocyte cell adhesion molecule a |
| chr25_-_31629095 | 1.56 |
ENSDART00000170673
ENSDART00000166930 |
lamb1a
|
laminin, beta 1a |
| chr3_+_23742868 | 1.54 |
ENSDART00000153512
|
hoxb3a
|
homeobox B3a |
| chr14_+_52563794 | 1.50 |
ENSDART00000168874
|
rpl26
|
ribosomal protein L26 |
| chr19_+_16222618 | 1.48 |
ENSDART00000137189
ENSDART00000169246 ENSDART00000190583 ENSDART00000189521 |
ptprua
|
protein tyrosine phosphatase, receptor type, U, a |
| chr10_+_9159279 | 1.46 |
ENSDART00000064968
|
rasgef1bb
|
RasGEF domain family, member 1Bb |
| chr20_+_34845672 | 1.42 |
ENSDART00000128895
|
emilin1a
|
elastin microfibril interfacer 1a |
| chr7_-_17028015 | 1.40 |
ENSDART00000022441
|
dbx1a
|
developing brain homeobox 1a |
| chr11_-_3343463 | 1.40 |
ENSDART00000066177
|
tuba2
|
tubulin, alpha 2 |
| chr4_-_16353733 | 1.40 |
ENSDART00000186785
|
lum
|
lumican |
| chr22_+_10752511 | 1.38 |
ENSDART00000081188
|
lsm7
|
LSM7 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr24_-_26304386 | 1.37 |
ENSDART00000175416
|
otos
|
otospiralin |
| chr21_-_37733287 | 1.35 |
ENSDART00000157826
|
mpp1
|
membrane protein, palmitoylated 1 |
| chr6_-_39275793 | 1.34 |
ENSDART00000180477
ENSDART00000148531 |
arhgef25b
|
Rho guanine nucleotide exchange factor (GEF) 25b |
| chr11_-_23219367 | 1.34 |
ENSDART00000003646
|
optc
|
opticin |
| chr2_+_25816843 | 1.34 |
ENSDART00000078639
|
slc2a2
|
solute carrier family 2 (facilitated glucose transporter), member 2 |
| chr20_-_34801181 | 1.34 |
ENSDART00000048375
ENSDART00000132426 |
stmn4
|
stathmin-like 4 |
| chr21_-_15674802 | 1.33 |
ENSDART00000136666
|
mmp11b
|
matrix metallopeptidase 11b |
| chr22_+_10752787 | 1.33 |
ENSDART00000186542
|
lsm7
|
LSM7 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr2_+_29976419 | 1.29 |
ENSDART00000056748
|
en2b
|
engrailed homeobox 2b |
| chr6_+_8176486 | 1.26 |
ENSDART00000193308
|
nfil3-5
|
nuclear factor, interleukin 3 regulated, member 5 |
| chr9_+_2002701 | 1.25 |
ENSDART00000082329
|
evx2
|
even-skipped homeobox 2 |
| chr6_-_31806345 | 1.25 |
ENSDART00000149369
|
si:dkey-209n16.2
|
si:dkey-209n16.2 |
| chr8_+_34731982 | 1.24 |
ENSDART00000066050
|
hpdb
|
4-hydroxyphenylpyruvate dioxygenase b |
| chr23_-_24682244 | 1.24 |
ENSDART00000104035
|
ctdsp2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr2_-_59145027 | 1.23 |
ENSDART00000128320
|
FO834803.1
|
|
| chr22_+_11535131 | 1.21 |
ENSDART00000113930
|
npb
|
neuropeptide B |
| chr13_+_29771463 | 1.21 |
ENSDART00000134424
ENSDART00000138332 ENSDART00000134330 ENSDART00000160944 ENSDART00000076992 ENSDART00000160921 |
pax2a
|
paired box 2a |
| chr8_-_38477817 | 1.21 |
ENSDART00000075989
|
inpp5l
|
inositol polyphosphate-5-phosphatase L |
| chr10_-_7988396 | 1.20 |
ENSDART00000141445
ENSDART00000024282 |
ewsr1a
|
EWS RNA-binding protein 1a |
| chr1_+_41588170 | 1.20 |
ENSDART00000139175
|
si:dkey-56e3.2
|
si:dkey-56e3.2 |
| chr6_-_34008827 | 1.19 |
ENSDART00000191183
ENSDART00000003701 ENSDART00000192502 |
tie1
|
tyrosine kinase with immunoglobulin-like and EGF-like domains 1 |
| chr8_+_20880848 | 1.18 |
ENSDART00000134488
ENSDART00000138605 ENSDART00000192234 |
si:ch73-196i15.3
|
si:ch73-196i15.3 |
| chr6_-_10964083 | 1.18 |
ENSDART00000181583
|
notum2
|
notum pectinacetylesterase 2 |
| chr24_+_28525507 | 1.18 |
ENSDART00000191121
|
arhgap29a
|
Rho GTPase activating protein 29a |
| chr17_-_42213822 | 1.17 |
ENSDART00000187904
ENSDART00000180029 |
nkx2.2a
|
NK2 homeobox 2a |
| chr24_+_23742690 | 1.17 |
ENSDART00000130162
|
TCF24
|
transcription factor 24 |
| chr16_+_11573471 | 1.17 |
ENSDART00000145858
|
si:dkey-11o1.2
|
si:dkey-11o1.2 |
| chr17_-_30702411 | 1.17 |
ENSDART00000114358
|
zgc:194392
|
zgc:194392 |
| chr10_+_8767541 | 1.16 |
ENSDART00000170272
|
itga1
|
integrin, alpha 1 |
| chr9_-_21904695 | 1.13 |
ENSDART00000134768
|
lmo7a
|
LIM domain 7a |
| chr23_+_9220436 | 1.13 |
ENSDART00000033663
ENSDART00000139870 |
rps21
|
ribosomal protein S21 |
| chr16_-_42013858 | 1.13 |
ENSDART00000045403
|
etv2
|
ets variant 2 |
| chr16_+_7662609 | 1.12 |
ENSDART00000184895
ENSDART00000149404 ENSDART00000081418 ENSDART00000081422 |
bves
|
blood vessel epicardial substance |
| chr8_+_39511932 | 1.12 |
ENSDART00000113511
|
lzts1
|
leucine zipper, putative tumor suppressor 1 |
| chr21_+_5209716 | 1.11 |
ENSDART00000102539
ENSDART00000053148 ENSDART00000102536 |
st8sia5
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5 |
| chr6_-_7438584 | 1.10 |
ENSDART00000053776
|
fkbp11
|
FK506 binding protein 11 |
| chr6_-_39765546 | 1.08 |
ENSDART00000185767
|
pfkmb
|
phosphofructokinase, muscle b |
| chr6_-_50203682 | 1.08 |
ENSDART00000083999
ENSDART00000143050 |
raly
|
RALY heterogeneous nuclear ribonucleoprotein |
| chr9_-_9348058 | 1.07 |
ENSDART00000132257
|
zgc:113337
|
zgc:113337 |
| chr12_+_638435 | 1.07 |
ENSDART00000152508
|
si:ch211-176g6.1
|
si:ch211-176g6.1 |
| chr1_-_21723329 | 1.07 |
ENSDART00000137138
|
si:ch211-134c9.2
|
si:ch211-134c9.2 |
| chr8_-_28349859 | 1.06 |
ENSDART00000062671
|
tuba8l
|
tubulin, alpha 8 like |
| chr5_+_51026563 | 1.05 |
ENSDART00000050988
|
gcnt4a
|
glucosaminyl (N-acetyl) transferase 4, core 2, a |
| chr5_+_4338874 | 1.05 |
ENSDART00000141866
|
sat1a.1
|
spermidine/spermine N1-acetyltransferase 1a, duplicate 1 |
| chr4_-_685412 | 1.04 |
ENSDART00000168167
|
rtcb
|
RNA 2',3'-cyclic phosphate and 5'-OH ligase |
| chr4_+_41227127 | 1.03 |
ENSDART00000136445
|
si:dkey-16p19.5
|
si:dkey-16p19.5 |
| chr10_+_22012389 | 1.03 |
ENSDART00000035188
|
kcnip1b
|
Kv channel interacting protein 1 b |
| chr3_+_32443395 | 1.03 |
ENSDART00000188447
|
prr12b
|
proline rich 12b |
| chr16_-_42014272 | 0.98 |
ENSDART00000180488
|
etv2
|
ets variant 2 |
| chr17_-_12926456 | 0.97 |
ENSDART00000044126
|
INSM2
|
INSM transcriptional repressor 2 |
| chr9_-_23922778 | 0.96 |
ENSDART00000135769
|
col6a3
|
collagen, type VI, alpha 3 |
| chr8_-_52715911 | 0.95 |
ENSDART00000168241
|
tubb2b
|
tubulin, beta 2b |
| chr23_-_32304650 | 0.95 |
ENSDART00000143772
ENSDART00000085642 ENSDART00000188989 |
dgkaa
|
diacylglycerol kinase, alpha a |
| chr25_-_12923482 | 0.94 |
ENSDART00000161754
|
CR450808.1
|
|
| chr18_+_8346920 | 0.94 |
ENSDART00000083421
|
cpt1b
|
carnitine palmitoyltransferase 1B (muscle) |
| chr6_-_10963887 | 0.93 |
ENSDART00000151102
|
notum2
|
notum pectinacetylesterase 2 |
| chr14_+_9462726 | 0.93 |
ENSDART00000063559
|
rell2
|
RELT-like 2 |
| chr18_-_21271373 | 0.91 |
ENSDART00000060001
|
pnp6
|
purine nucleoside phosphorylase 6 |
| chr4_+_25220674 | 0.90 |
ENSDART00000066934
|
itih5
|
inter-alpha-trypsin inhibitor heavy chain family, member 5 |
| chr20_+_48803248 | 0.88 |
ENSDART00000164006
|
nkx2.4b
|
NK2 homeobox 4b |
| chr16_+_12836143 | 0.88 |
ENSDART00000067741
|
cacng6b
|
calcium channel, voltage-dependent, gamma subunit 6b |
| chr15_-_8517555 | 0.87 |
ENSDART00000140213
|
npas1
|
neuronal PAS domain protein 1 |
| chr4_+_27131725 | 0.87 |
ENSDART00000013083
|
brd1a
|
bromodomain containing 1a |
| chr25_+_25438165 | 0.86 |
ENSDART00000161369
|
LRRC56
|
si:ch211-103e16.5 |
| chr20_+_1996202 | 0.86 |
ENSDART00000184143
|
CABZ01092781.1
|
|
| chr7_-_38340674 | 0.84 |
ENSDART00000075782
|
slc7a10a
|
solute carrier family 7 (neutral amino acid transporter light chain, asc system), member 10a |
| chr9_-_1986014 | 0.84 |
ENSDART00000142842
|
hoxd12a
|
homeobox D12a |
| chr21_+_41743493 | 0.82 |
ENSDART00000192669
|
ppp2r2bb
|
protein phosphatase 2, regulatory subunit B, beta b |
| chr2_-_10188598 | 0.81 |
ENSDART00000189122
|
dmbx1a
|
diencephalon/mesencephalon homeobox 1a |
| chr19_+_19511394 | 0.80 |
ENSDART00000172410
|
jazf1a
|
JAZF zinc finger 1a |
| chr8_+_23485079 | 0.80 |
ENSDART00000026316
|
sema3gb
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Gb |
| chr8_+_25079470 | 0.80 |
ENSDART00000000744
|
sypl2b
|
synaptophysin-like 2b |
| chr4_-_27398385 | 0.78 |
ENSDART00000142117
ENSDART00000150553 ENSDART00000182746 |
fam19a5a
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5a |
| chr7_+_44445595 | 0.78 |
ENSDART00000108766
|
cdh5
|
cadherin 5 |
| chr23_+_42304602 | 0.77 |
ENSDART00000166004
|
cyp2aa11
|
cytochrome P450, family 2, subfamily AA, polypeptide 11 |
| chr8_+_27743550 | 0.77 |
ENSDART00000046004
|
wnt2bb
|
wingless-type MMTV integration site family, member 2Bb |
| chr19_-_42557416 | 0.76 |
ENSDART00000163217
ENSDART00000128278 ENSDART00000162304 ENSDART00000166556 |
si:dkey-267n13.1
|
si:dkey-267n13.1 |
| chr23_-_30781875 | 0.76 |
ENSDART00000114628
ENSDART00000180949 ENSDART00000191313 |
myt1a
|
myelin transcription factor 1a |
| chr24_+_25471196 | 0.74 |
ENSDART00000066625
|
smpx
|
small muscle protein, X-linked |
| chr7_+_24114694 | 0.74 |
ENSDART00000127177
|
mrpl52
|
mitochondrial ribosomal protein L52 |
| chr21_-_12119711 | 0.73 |
ENSDART00000131538
|
celf4
|
CUGBP, Elav-like family member 4 |
| chr20_-_45807982 | 0.73 |
ENSDART00000074546
|
fermt1
|
fermitin family member 1 |
| chr9_+_30720048 | 0.72 |
ENSDART00000146115
|
klf12b
|
Kruppel-like factor 12b |
| chr10_+_34394454 | 0.72 |
ENSDART00000110121
|
stard13a
|
StAR-related lipid transfer (START) domain containing 13a |
| chr7_+_36552725 | 0.72 |
ENSDART00000173682
|
chd9
|
chromodomain helicase DNA binding protein 9 |
| chr2_-_43168292 | 0.72 |
ENSDART00000132588
|
crema
|
cAMP responsive element modulator a |
| chr9_+_10014817 | 0.71 |
ENSDART00000132065
|
nxph2a
|
neurexophilin 2a |
| chr22_-_17256573 | 0.70 |
ENSDART00000136119
ENSDART00000062891 |
nphs2
|
nephrosis 2, idiopathic, steroid-resistant (podocin) |
| chr16_+_19014886 | 0.70 |
ENSDART00000079298
|
si:ch211-254p10.2
|
si:ch211-254p10.2 |
| chr6_-_40697585 | 0.69 |
ENSDART00000113196
|
si:ch211-157b11.14
|
si:ch211-157b11.14 |
| chr1_+_22002649 | 0.69 |
ENSDART00000141317
|
dnah6
|
dynein, axonemal, heavy chain 6 |
| chr22_-_29242347 | 0.68 |
ENSDART00000040761
|
pvalb7
|
parvalbumin 7 |
| chr22_-_3152357 | 0.68 |
ENSDART00000170983
|
lmnb2
|
lamin B2 |
| chr15_+_7054754 | 0.68 |
ENSDART00000149800
|
foxl2a
|
forkhead box L2a |
| chr21_-_22317920 | 0.67 |
ENSDART00000191083
ENSDART00000108701 |
gdpd4b
|
glycerophosphodiester phosphodiesterase domain containing 4b |
| chr19_+_37620342 | 0.66 |
ENSDART00000158960
|
thsd7aa
|
thrombospondin, type I, domain containing 7Aa |
| chr7_-_59054322 | 0.66 |
ENSDART00000165390
|
chmp5b
|
charged multivesicular body protein 5b |
| chr9_-_23990416 | 0.64 |
ENSDART00000113176
|
col6a3
|
collagen, type VI, alpha 3 |
| chr6_-_46861676 | 0.63 |
ENSDART00000188712
ENSDART00000190148 |
igfn1.3
|
immunoglobulin-like and fibronectin type III domain containing 1, tandem duplicate 3 |
| chr7_+_35238234 | 0.63 |
ENSDART00000178732
|
tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr21_-_40799404 | 0.63 |
ENSDART00000147405
ENSDART00000157512 |
zgc:162239
|
zgc:162239 |
| chr16_+_10972733 | 0.63 |
ENSDART00000049323
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr11_-_3865472 | 0.61 |
ENSDART00000161426
|
gata2a
|
GATA binding protein 2a |
| chr23_-_45705525 | 0.60 |
ENSDART00000148959
|
ednrab
|
endothelin receptor type Ab |
| chr5_-_5147041 | 0.60 |
ENSDART00000180236
|
lmx1ba
|
LIM homeobox transcription factor 1, beta a |
| chr3_-_18792492 | 0.60 |
ENSDART00000134208
ENSDART00000034373 |
hagh
|
hydroxyacylglutathione hydrolase |
| chr3_+_24634481 | 0.60 |
ENSDART00000163080
|
si:dkey-68o6.8
|
si:dkey-68o6.8 |
| chr1_+_33383971 | 0.60 |
ENSDART00000150043
|
dhrsx
|
dehydrogenase/reductase (SDR family) X-linked |
| chr1_+_31110817 | 0.59 |
ENSDART00000137863
|
eef1a1b
|
eukaryotic translation elongation factor 1 alpha 1b |
| chr12_-_7854216 | 0.59 |
ENSDART00000149594
|
ank3b
|
ankyrin 3b |
| chr9_+_10014514 | 0.59 |
ENSDART00000185590
|
nxph2a
|
neurexophilin 2a |
| chr9_-_19161982 | 0.59 |
ENSDART00000081878
|
pou1f1
|
POU class 1 homeobox 1 |
| chr3_-_22228602 | 0.58 |
ENSDART00000017750
|
myl4
|
myosin, light chain 4, alkali; atrial, embryonic |
| chr15_+_6652396 | 0.58 |
ENSDART00000192813
ENSDART00000157678 |
nop53
|
NOP53 ribosome biogenesis factor |
| chr12_-_45197038 | 0.57 |
ENSDART00000016635
|
bccip
|
BRCA2 and CDKN1A interacting protein |
| chr22_-_15280638 | 0.57 |
ENSDART00000063008
|
mfng
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr8_+_1187928 | 0.57 |
ENSDART00000127252
|
slc35d2
|
solute carrier family 35 (UDP-GlcNAc/UDP-glucose transporter), member D2 |
| chr2_-_51794472 | 0.56 |
ENSDART00000186652
|
BX908782.3
|
|
| chr9_+_1138323 | 0.54 |
ENSDART00000190352
ENSDART00000190387 |
slc15a1a
|
solute carrier family 15 (oligopeptide transporter), member 1a |
| chr25_+_24616717 | 0.54 |
ENSDART00000089113
|
abtb2b
|
ankyrin repeat and BTB (POZ) domain containing 2b |
| chr14_-_3070613 | 0.54 |
ENSDART00000193729
ENSDART00000090196 |
slc35a4
|
solute carrier family 35, member A4 |
| chr23_+_35969228 | 0.53 |
ENSDART00000179992
ENSDART00000184107 |
hoxc10a
|
homeobox C10a |
| chr15_-_5172170 | 0.52 |
ENSDART00000062830
|
or126-5
|
odorant receptor, family E, subfamily 126, member 5 |
| chr19_+_27342479 | 0.52 |
ENSDART00000184687
|
ddx39b
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39B |
| chr15_-_8517376 | 0.52 |
ENSDART00000186289
|
npas1
|
neuronal PAS domain protein 1 |
| chr13_+_3252950 | 0.52 |
ENSDART00000020671
|
prph2b
|
peripherin 2b (retinal degeneration, slow) |
| chr11_-_37509001 | 0.50 |
ENSDART00000109753
|
bsnb
|
bassoon (presynaptic cytomatrix protein) b |
| chr18_-_17485419 | 0.50 |
ENSDART00000018764
|
foxl1
|
forkhead box L1 |
| chr13_-_12005429 | 0.49 |
ENSDART00000180302
|
mgea5
|
meningioma expressed antigen 5 (hyaluronidase) |
| chr10_-_5581487 | 0.49 |
ENSDART00000141943
|
syk
|
spleen tyrosine kinase |
| chr2_-_1548330 | 0.49 |
ENSDART00000082155
ENSDART00000108481 ENSDART00000111272 |
dab1b
|
Dab, reelin signal transducer, homolog 1b (Drosophila) |
| chr25_+_25438322 | 0.48 |
ENSDART00000150364
|
LRRC56
|
si:ch211-103e16.5 |
| chr14_-_32089117 | 0.48 |
ENSDART00000158014
|
si:ch211-69b22.5
|
si:ch211-69b22.5 |
| chr5_-_10946232 | 0.48 |
ENSDART00000163139
ENSDART00000031265 |
rtn4r
|
reticulon 4 receptor |
| chr2_+_20410652 | 0.48 |
ENSDART00000185940
|
palmda
|
palmdelphin a |
| chr3_-_16289826 | 0.48 |
ENSDART00000131972
|
bckdhbl
|
branched chain keto acid dehydrogenase E1, beta polypeptide, like |
| chr14_+_22591624 | 0.47 |
ENSDART00000108987
|
gfra4b
|
GDNF family receptor alpha 4b |
| chr16_-_14353567 | 0.47 |
ENSDART00000139859
|
itga10
|
integrin, alpha 10 |
| chr5_+_20421539 | 0.46 |
ENSDART00000164499
|
selplg
|
selectin P ligand |
| chr18_-_28938912 | 0.46 |
ENSDART00000136201
|
si:ch211-174j14.2
|
si:ch211-174j14.2 |
| chr4_-_12104421 | 0.46 |
ENSDART00000139561
|
mrps33
|
mitochondrial ribosomal protein S33 |
| chr8_-_53680653 | 0.46 |
ENSDART00000164739
|
wnt5a
|
wingless-type MMTV integration site family, member 5a |
| chr21_-_30658509 | 0.45 |
ENSDART00000139764
|
si:dkey-22f5.9
|
si:dkey-22f5.9 |
| chr6_-_15101477 | 0.44 |
ENSDART00000187713
ENSDART00000124132 |
fhl2b
|
four and a half LIM domains 2b |
| chr25_+_26844028 | 0.43 |
ENSDART00000127274
ENSDART00000156179 |
sema7a
|
semaphorin 7A |
| chr17_+_29348013 | 0.42 |
ENSDART00000188885
ENSDART00000192005 ENSDART00000190484 ENSDART00000188565 |
kctd3
|
potassium channel tetramerization domain containing 3 |
| chr8_+_2487250 | 0.42 |
ENSDART00000081325
|
dynll1
|
dynein, light chain, LC8-type 1 |
| chr16_+_14029283 | 0.41 |
ENSDART00000146165
ENSDART00000132075 |
rusc1
|
RUN and SH3 domain containing 1 |
| chr13_-_39830999 | 0.40 |
ENSDART00000115089
|
zgc:171482
|
zgc:171482 |
| chr4_+_63769987 | 0.40 |
ENSDART00000168878
|
znf1048
|
zinc finger protein 1048 |
| chr5_-_5147220 | 0.40 |
ENSDART00000187026
ENSDART00000162334 |
lmx1ba
|
LIM homeobox transcription factor 1, beta a |
Network of associatons between targets according to the STRING database.
First level regulatory network of mecom
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.8 | 3.1 | GO:0055130 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.7 | 3.7 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.5 | 2.1 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.5 | 2.8 | GO:0035176 | mammillary body development(GO:0021767) social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.4 | 1.7 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.4 | 2.5 | GO:0021885 | forebrain cell migration(GO:0021885) |
| 0.4 | 1.2 | GO:0021530 | spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.4 | 2.0 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.4 | 2.3 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.3 | 2.7 | GO:0018904 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.3 | 1.3 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.3 | 0.9 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.3 | 1.8 | GO:0090133 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.3 | 1.2 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.3 | 2.3 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.3 | 0.8 | GO:0042941 | D-amino acid transport(GO:0042940) D-alanine transport(GO:0042941) D-serine transport(GO:0042942) |
| 0.3 | 1.0 | GO:0009447 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.3 | 1.6 | GO:0055016 | hypochord development(GO:0055016) |
| 0.2 | 3.2 | GO:0042396 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.2 | 1.6 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.2 | 0.7 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.2 | 2.0 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) |
| 0.2 | 1.2 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.2 | 0.6 | GO:1902893 | pri-miRNA transcription from RNA polymerase II promoter(GO:0061614) regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902893) positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.2 | 1.2 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.2 | 0.6 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.2 | 0.8 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.2 | 1.1 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.2 | 0.9 | GO:1902514 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.2 | 0.5 | GO:0045579 | positive regulation of B cell differentiation(GO:0045579) |
| 0.2 | 2.4 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.2 | 1.4 | GO:0070293 | renal absorption(GO:0070293) |
| 0.1 | 0.4 | GO:0060907 | macrophage cytokine production(GO:0010934) regulation of macrophage cytokine production(GO:0010935) positive regulation of macrophage cytokine production(GO:0060907) |
| 0.1 | 5.0 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 6.9 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 0.8 | GO:0001955 | blood vessel maturation(GO:0001955) regulation of establishment of cell polarity(GO:2000114) |
| 0.1 | 1.3 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 | 0.6 | GO:0003319 | cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) |
| 0.1 | 2.2 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.1 | 3.7 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.1 | 1.0 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 1.8 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.1 | 1.1 | GO:0006735 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 0.7 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.1 | 1.0 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.1 | 0.8 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 0.2 | GO:0071314 | response to cocaine(GO:0042220) cellular response to alkaloid(GO:0071312) cellular response to cocaine(GO:0071314) |
| 0.1 | 0.5 | GO:0097241 | hematopoietic stem cell migration to bone marrow(GO:0097241) |
| 0.1 | 1.2 | GO:0048016 | inositol phosphate-mediated signaling(GO:0048016) |
| 0.1 | 0.5 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.1 | 0.9 | GO:0030878 | thyroid gland development(GO:0030878) |
| 0.1 | 0.6 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.1 | 0.4 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.1 | 0.9 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.1 | 0.2 | GO:0070257 | regulation of mucus secretion(GO:0070255) positive regulation of mucus secretion(GO:0070257) |
| 0.1 | 1.8 | GO:0031629 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.1 | 2.4 | GO:0030901 | midbrain development(GO:0030901) |
| 0.1 | 0.9 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.0 | 0.9 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.5 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.0 | 0.1 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.0 | 1.1 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 1.0 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 1.2 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.0 | 1.4 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.9 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.5 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.4 | GO:1900407 | regulation of cellular response to oxidative stress(GO:1900407) |
| 0.0 | 1.1 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.0 | 0.3 | GO:0099558 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.0 | 4.0 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 0.7 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 0.5 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 1.3 | GO:1902749 | regulation of cell cycle G2/M phase transition(GO:1902749) |
| 0.0 | 2.5 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 2.4 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 1.3 | GO:0021515 | cell differentiation in spinal cord(GO:0021515) |
| 0.0 | 0.1 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.6 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 1.6 | GO:0010770 | positive regulation of cell morphogenesis involved in differentiation(GO:0010770) |
| 0.0 | 1.1 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.7 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 1.6 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 2.6 | GO:0006979 | response to oxidative stress(GO:0006979) |
| 0.0 | 0.7 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.1 | GO:0018199 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 0.7 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 1.4 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 1.8 | GO:0021782 | glial cell development(GO:0021782) |
| 0.0 | 2.5 | GO:0048793 | pronephros development(GO:0048793) |
| 0.0 | 0.6 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.8 | GO:0017144 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.5 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.2 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.3 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 0.0 | 0.1 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.0 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.7 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.9 | 3.6 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.4 | 2.7 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.3 | 4.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.2 | 2.2 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.2 | 6.9 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.7 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 1.0 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 3.5 | GO:0046930 | pore complex(GO:0046930) |
| 0.1 | 0.9 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 4.9 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 1.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 1.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 1.0 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.1 | 1.8 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 1.6 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 0.5 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.0 | 0.9 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.6 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 1.6 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.3 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 3.1 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.4 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.5 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.3 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.7 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.6 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 1.8 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.1 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 0.8 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.3 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic cytoskeleton(GO:0099569) |
| 0.0 | 4.1 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 1.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.3 | GO:0032589 | neuron projection membrane(GO:0032589) dendrite membrane(GO:0032590) |
| 0.0 | 12.7 | GO:0005615 | extracellular space(GO:0005615) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.5 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.8 | 3.1 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.6 | 1.8 | GO:0031704 | apelin receptor binding(GO:0031704) |
| 0.5 | 2.1 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.5 | 2.6 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.4 | 1.7 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.4 | 1.2 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.3 | 2.4 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.2 | 3.2 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.2 | 3.7 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.2 | 1.0 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.2 | 1.9 | GO:0015250 | water channel activity(GO:0015250) |
| 0.2 | 1.3 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 1.0 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.1 | 0.6 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.6 | GO:0005461 | UDP-glucuronic acid transmembrane transporter activity(GO:0005461) UDP-N-acetylgalactosamine transmembrane transporter activity(GO:0005463) |
| 0.1 | 1.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 1.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 1.2 | GO:0052659 | inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.1 | 0.9 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 0.6 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 2.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 5.6 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 0.9 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 1.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 2.7 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 0.6 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.1 | 2.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 0.6 | GO:0071916 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.1 | 0.2 | GO:0016496 | substance P receptor activity(GO:0016496) |
| 0.1 | 0.7 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.1 | 1.1 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 0.9 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 2.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 2.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.8 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) structural constituent of synapse(GO:0098918) |
| 0.0 | 0.6 | GO:0031729 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.0 | 0.9 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 4.0 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.5 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.9 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 1.2 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.0 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.5 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.2 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 1.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.7 | GO:0042562 | hormone binding(GO:0042562) |
| 0.0 | 2.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 5.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 1.1 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.3 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 2.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.0 | 0.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.2 | GO:0030955 | potassium ion binding(GO:0030955) alkali metal ion binding(GO:0031420) |
| 0.0 | 1.2 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.8 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.8 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.0 | 0.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.6 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.0 | 1.2 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.2 | 3.7 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 5.2 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 2.5 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 2.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 1.6 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.1 | 0.8 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.1 | 2.3 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.5 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.5 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 3.7 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.7 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.5 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.3 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.7 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 2.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.5 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.6 | PID P73PATHWAY | p73 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 6.4 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.3 | 2.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.3 | 2.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.2 | 2.9 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.2 | 1.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 5.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 1.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 1.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 1.1 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 0.9 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 1.0 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.3 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 0.4 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.6 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 1.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.5 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 2.6 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.6 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.1 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |