Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
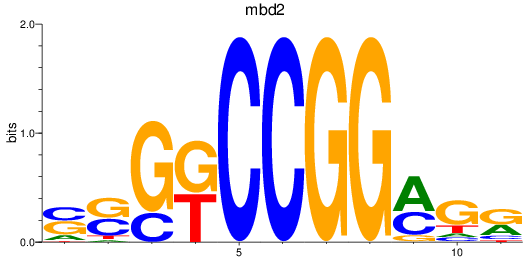
Results for mbd2
Z-value: 0.46
Transcription factors associated with mbd2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mbd2
|
ENSDARG00000075952 | methyl-CpG binding domain protein 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mbd2 | dr11_v1_chr5_-_1047222_1047222 | 0.66 | 5.3e-02 | Click! |
Activity profile of mbd2 motif
Sorted Z-values of mbd2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_24251057 | 0.21 |
ENSDART00000114169
|
bnip1a
|
BCL2 interacting protein 1a |
| chr20_+_16743056 | 0.19 |
ENSDART00000050308
|
calm1b
|
calmodulin 1b |
| chr20_-_40717900 | 0.18 |
ENSDART00000181663
|
cx43
|
connexin 43 |
| chr16_-_45058919 | 0.16 |
ENSDART00000177134
|
gapdhs
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr23_-_3681026 | 0.16 |
ENSDART00000192128
ENSDART00000040086 |
pacsin1a
|
protein kinase C and casein kinase substrate in neurons 1a |
| chr12_+_27462225 | 0.15 |
ENSDART00000105661
|
meox1
|
mesenchyme homeobox 1 |
| chr23_+_21964325 | 0.14 |
ENSDART00000140208
ENSDART00000135690 |
lactbl1a
|
lactamase, beta-like 1a |
| chr4_+_16885854 | 0.14 |
ENSDART00000017726
|
etnk1
|
ethanolamine kinase 1 |
| chr9_+_53725294 | 0.14 |
ENSDART00000165991
|
cnmd
|
chondromodulin |
| chr9_+_53725128 | 0.13 |
ENSDART00000169062
|
cnmd
|
chondromodulin |
| chr18_+_62932 | 0.13 |
ENSDART00000052638
|
slc27a2a
|
solute carrier family 27 (fatty acid transporter), member 2a |
| chr16_+_20915319 | 0.13 |
ENSDART00000079383
|
hoxa9b
|
homeobox A9b |
| chr18_-_46464501 | 0.12 |
ENSDART00000040669
|
sphkap
|
SPHK1 interactor, AKAP domain containing |
| chr8_-_54223316 | 0.12 |
ENSDART00000018054
|
trh
|
thyrotropin-releasing hormone |
| chr23_+_45584223 | 0.12 |
ENSDART00000149367
|
si:ch73-290k24.5
|
si:ch73-290k24.5 |
| chr8_+_54013199 | 0.12 |
ENSDART00000158497
|
CABZ01079663.1
|
|
| chr18_+_54354 | 0.12 |
ENSDART00000097163
|
zgc:158482
|
zgc:158482 |
| chr4_-_24019711 | 0.11 |
ENSDART00000077926
|
celf2
|
cugbp, Elav-like family member 2 |
| chr11_-_762721 | 0.11 |
ENSDART00000166465
|
syn2b
|
synapsin IIb |
| chr22_-_12862415 | 0.11 |
ENSDART00000145156
ENSDART00000137280 |
glsa
|
glutaminase a |
| chr1_-_30689004 | 0.11 |
ENSDART00000018827
|
dachc
|
dachshund c |
| chr21_+_26697536 | 0.11 |
ENSDART00000004109
|
gng3
|
guanine nucleotide binding protein (G protein), gamma 3 |
| chr4_-_77979432 | 0.09 |
ENSDART00000049170
|
zgc:85975
|
zgc:85975 |
| chr1_+_32521469 | 0.09 |
ENSDART00000113818
ENSDART00000152580 |
nlgn4a
|
neuroligin 4a |
| chr22_-_3564563 | 0.09 |
ENSDART00000145114
|
ptprsa
|
protein tyrosine phosphatase, receptor type, s, a |
| chr13_-_1349922 | 0.09 |
ENSDART00000140970
|
si:ch73-52p7.1
|
si:ch73-52p7.1 |
| chr9_-_21936841 | 0.09 |
ENSDART00000144843
|
lmo7a
|
LIM domain 7a |
| chr25_+_21833287 | 0.09 |
ENSDART00000187606
|
ckmt1
|
creatine kinase, mitochondrial 1 |
| chr16_+_5678071 | 0.09 |
ENSDART00000011166
ENSDART00000134198 ENSDART00000131575 |
zgc:158689
|
zgc:158689 |
| chr3_-_25813426 | 0.09 |
ENSDART00000039482
|
ntn1b
|
netrin 1b |
| chr21_-_12272543 | 0.08 |
ENSDART00000081510
ENSDART00000151297 |
celf4
|
CUGBP, Elav-like family member 4 |
| chr22_+_110158 | 0.08 |
ENSDART00000143698
|
prkar2ab
|
protein kinase, cAMP-dependent, regulatory, type II, alpha, B |
| chr7_+_30867008 | 0.08 |
ENSDART00000193106
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr6_+_39222598 | 0.08 |
ENSDART00000154991
|
b4galnt1b
|
beta-1,4-N-acetyl-galactosaminyl transferase 1b |
| chr12_+_25600685 | 0.08 |
ENSDART00000077157
|
six3b
|
SIX homeobox 3b |
| chr15_+_30158652 | 0.08 |
ENSDART00000190682
|
nlk2
|
nemo-like kinase, type 2 |
| chr22_+_9060699 | 0.07 |
ENSDART00000133993
|
BX248395.1
|
|
| chr19_-_27966526 | 0.07 |
ENSDART00000141896
|
ube2ql1
|
ubiquitin-conjugating enzyme E2Q family-like 1 |
| chr8_+_2878756 | 0.07 |
ENSDART00000168107
|
crb2b
|
crumbs family member 2b |
| chr5_-_34616599 | 0.07 |
ENSDART00000050271
ENSDART00000097975 |
hexb
|
hexosaminidase B (beta polypeptide) |
| chr13_-_51922290 | 0.07 |
ENSDART00000168648
|
srfb
|
serum response factor b |
| chr20_+_54738210 | 0.07 |
ENSDART00000151399
|
pak7
|
p21 protein (Cdc42/Rac)-activated kinase 7 |
| chr17_-_6382392 | 0.07 |
ENSDART00000188051
ENSDART00000192560 ENSDART00000137389 ENSDART00000115389 |
txlnbb
|
taxilin beta b |
| chr8_+_25900049 | 0.06 |
ENSDART00000124300
ENSDART00000127618 ENSDART00000024009 |
rhoab
|
ras homolog gene family, member Ab |
| chr7_+_53755054 | 0.06 |
ENSDART00000181629
|
neo1a
|
neogenin 1a |
| chr7_+_53754653 | 0.06 |
ENSDART00000163261
ENSDART00000158160 |
neo1a
|
neogenin 1a |
| chr13_-_27620815 | 0.06 |
ENSDART00000139904
|
kcnq5a
|
potassium voltage-gated channel, KQT-like subfamily, member 5a |
| chr9_-_5046315 | 0.06 |
ENSDART00000179087
ENSDART00000109954 |
nr4a2a
|
nuclear receptor subfamily 4, group A, member 2a |
| chr2_+_56139941 | 0.06 |
ENSDART00000164741
|
pgpep1
|
pyroglutamyl-peptidase I |
| chr13_-_1408775 | 0.06 |
ENSDART00000049684
|
bag2
|
BCL2 associated athanogene 2 |
| chr21_+_22833905 | 0.06 |
ENSDART00000111150
|
birc2
|
baculoviral IAP repeat containing 2 |
| chr9_+_44034790 | 0.06 |
ENSDART00000166110
ENSDART00000176954 |
itga4
|
integrin alpha 4 |
| chr17_+_53250802 | 0.06 |
ENSDART00000143819
|
VASH1
|
vasohibin 1 |
| chr14_-_451555 | 0.06 |
ENSDART00000190906
|
FAT4
|
FAT atypical cadherin 4 |
| chr19_-_47276297 | 0.06 |
ENSDART00000141437
|
sdc2
|
syndecan 2 |
| chr14_-_3032016 | 0.05 |
ENSDART00000183461
ENSDART00000183035 |
ndst1a
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1a |
| chr7_-_35314347 | 0.05 |
ENSDART00000005053
|
slc12a4
|
solute carrier family 12 (potassium/chloride transporter), member 4 |
| chr24_-_35699595 | 0.05 |
ENSDART00000167990
|
mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr6_-_1874664 | 0.05 |
ENSDART00000007972
|
dlgap4b
|
discs, large (Drosophila) homolog-associated protein 4b |
| chr1_-_69444 | 0.05 |
ENSDART00000166954
|
si:zfos-1011f11.1
|
si:zfos-1011f11.1 |
| chr11_+_575665 | 0.05 |
ENSDART00000122133
|
mkrn2os.1
|
MKRN2 opposite strand, tandem duplicate 1 |
| chr19_-_27966780 | 0.05 |
ENSDART00000110016
|
ube2ql1
|
ubiquitin-conjugating enzyme E2Q family-like 1 |
| chr2_-_56635744 | 0.05 |
ENSDART00000167790
ENSDART00000168160 |
pip5k1cb
|
phosphatidylinositol-4-phosphate 5-kinase, type I, gamma b |
| chr16_-_27161410 | 0.05 |
ENSDART00000177503
|
LO017771.1
|
|
| chr18_-_46763170 | 0.05 |
ENSDART00000171880
|
dner
|
delta/notch-like EGF repeat containing |
| chr21_-_5393125 | 0.05 |
ENSDART00000146061
|
psmd5
|
proteasome 26S subunit, non-ATPase 5 |
| chr3_+_26081343 | 0.05 |
ENSDART00000134647
|
atp2a1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr13_-_45155284 | 0.05 |
ENSDART00000161144
|
runx3
|
runt-related transcription factor 3 |
| chr10_+_16225870 | 0.05 |
ENSDART00000164647
|
slc12a2
|
solute carrier family 12 (sodium/potassium/chloride transporter), member 2 |
| chr14_-_3031810 | 0.05 |
ENSDART00000090213
|
ndst1a
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1a |
| chr12_-_3756405 | 0.05 |
ENSDART00000150839
|
fam57bb
|
family with sequence similarity 57, member Bb |
| chr12_-_11457625 | 0.05 |
ENSDART00000012318
|
htra1b
|
HtrA serine peptidase 1b |
| chr19_+_5480327 | 0.05 |
ENSDART00000148794
|
jupb
|
junction plakoglobin b |
| chr8_+_1148876 | 0.04 |
ENSDART00000148651
|
avp
|
arginine vasopressin |
| chr19_-_45534392 | 0.04 |
ENSDART00000163920
|
trps1
|
trichorhinophalangeal syndrome I |
| chr20_+_40457599 | 0.04 |
ENSDART00000017553
|
serinc1
|
serine incorporator 1 |
| chr9_+_44034990 | 0.04 |
ENSDART00000162406
|
itga4
|
integrin alpha 4 |
| chr10_-_26999482 | 0.04 |
ENSDART00000146085
|
si:dkey-88p24.11
|
si:dkey-88p24.11 |
| chr12_-_3773869 | 0.04 |
ENSDART00000092983
|
si:ch211-166g5.4
|
si:ch211-166g5.4 |
| chr3_-_5228137 | 0.04 |
ENSDART00000137105
|
myh9b
|
myosin, heavy chain 9b, non-muscle |
| chr21_+_28728030 | 0.04 |
ENSDART00000097307
|
puraa
|
purine-rich element binding protein Aa |
| chr23_+_39331216 | 0.04 |
ENSDART00000160957
|
kcng1
|
potassium voltage-gated channel, subfamily G, member 1 |
| chr17_-_8673278 | 0.04 |
ENSDART00000171850
ENSDART00000017337 ENSDART00000148504 ENSDART00000148808 |
ctbp2a
|
C-terminal binding protein 2a |
| chr2_+_41876595 | 0.04 |
ENSDART00000112243
|
crlf1a
|
cytokine receptor-like factor 1a |
| chr12_+_5708400 | 0.03 |
ENSDART00000017191
|
dlx3b
|
distal-less homeobox 3b |
| chr10_-_28193642 | 0.03 |
ENSDART00000019050
|
rps6kb1a
|
ribosomal protein S6 kinase b, polypeptide 1a |
| chr18_+_3579829 | 0.03 |
ENSDART00000158763
ENSDART00000182850 ENSDART00000162754 ENSDART00000178789 ENSDART00000172656 |
lrch3
|
leucine-rich repeats and calponin homology (CH) domain containing 3 |
| chr14_+_44860335 | 0.03 |
ENSDART00000091620
ENSDART00000173043 ENSDART00000091625 |
atp8a1
|
ATPase phospholipid transporting 8A1 |
| chr5_+_71999996 | 0.03 |
ENSDART00000179933
ENSDART00000187070 |
PLPP7 (1 of many)
|
phospholipid phosphatase 7 (inactive) |
| chr2_+_35854242 | 0.03 |
ENSDART00000134918
|
dhx9
|
DEAH (Asp-Glu-Ala-His) box helicase 9 |
| chr23_-_17450746 | 0.03 |
ENSDART00000145399
ENSDART00000136457 ENSDART00000133125 ENSDART00000145719 ENSDART00000147524 ENSDART00000005366 ENSDART00000104680 |
tpd52l2b
|
tumor protein D52-like 2b |
| chr12_-_11294979 | 0.03 |
ENSDART00000148850
|
get4
|
golgi to ER traffic protein 4 homolog (S. cerevisiae) |
| chr1_-_22803147 | 0.03 |
ENSDART00000086867
|
tapt1b
|
transmembrane anterior posterior transformation 1b |
| chr20_+_18943406 | 0.03 |
ENSDART00000193590
|
mtmr9
|
myotubularin related protein 9 |
| chr4_+_11439511 | 0.03 |
ENSDART00000150485
|
pcloa
|
piccolo presynaptic cytomatrix protein a |
| chr15_-_3078600 | 0.03 |
ENSDART00000172842
|
fndc3a
|
fibronectin type III domain containing 3A |
| chr18_-_51015718 | 0.03 |
ENSDART00000190698
|
LO018598.1
|
|
| chr15_+_404891 | 0.03 |
ENSDART00000155682
|
nipsnap2
|
nipsnap homolog 2 |
| chr2_-_689047 | 0.03 |
ENSDART00000122732
|
foxc1a
|
forkhead box C1a |
| chr2_-_5399437 | 0.03 |
ENSDART00000132411
|
si:ch1073-184j22.2
|
si:ch1073-184j22.2 |
| chr12_+_763051 | 0.02 |
ENSDART00000152579
|
abcc3
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
| chr20_+_46699021 | 0.02 |
ENSDART00000167398
ENSDART00000029894 |
wdr26b
|
WD repeat domain 26b |
| chr15_+_36966369 | 0.02 |
ENSDART00000163622
|
kirrel3l
|
kirre like nephrin family adhesion molecule 3, like |
| chr24_+_13735616 | 0.02 |
ENSDART00000184267
|
msc
|
musculin (activated B-cell factor-1) |
| chr14_+_743346 | 0.02 |
ENSDART00000110511
|
klb
|
klotho beta |
| chr24_-_35699444 | 0.02 |
ENSDART00000166567
|
mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr24_-_21404367 | 0.02 |
ENSDART00000152093
|
atp8a2
|
ATPase phospholipid transporting 8A2 |
| chr11_+_44579865 | 0.02 |
ENSDART00000173425
|
nid1b
|
nidogen 1b |
| chr19_+_56351 | 0.02 |
ENSDART00000168334
|
col14a1b
|
collagen, type XIV, alpha 1b |
| chr13_+_4888351 | 0.02 |
ENSDART00000145940
|
micu1
|
mitochondrial calcium uptake 1 |
| chr7_-_2090594 | 0.02 |
ENSDART00000183166
|
si:cabz01007802.1
|
si:cabz01007802.1 |
| chr16_-_42238120 | 0.02 |
ENSDART00000084730
|
zgc:162160
|
zgc:162160 |
| chr7_-_32980017 | 0.01 |
ENSDART00000113744
|
pkp3b
|
plakophilin 3b |
| chr17_-_26604549 | 0.01 |
ENSDART00000174773
|
fam149b1
|
family with sequence similarity 149, member B1 |
| chr22_+_2111120 | 0.01 |
ENSDART00000165525
|
znf1144
|
zinc finger protein 1144 |
| chr21_+_1143141 | 0.01 |
ENSDART00000178294
|
CABZ01086139.1
|
|
| chr4_+_77933084 | 0.01 |
ENSDART00000148728
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr11_-_27738489 | 0.01 |
ENSDART00000181612
|
fam120a
|
family with sequence similarity 120A |
| chr15_-_3940671 | 0.01 |
ENSDART00000189741
|
mindy4b
|
MINDY family member 4B |
| chr13_-_40120252 | 0.01 |
ENSDART00000157852
|
crtac1b
|
cartilage acidic protein 1b |
| chr9_+_24159725 | 0.01 |
ENSDART00000137756
|
hecw2a
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2a |
| chr11_-_575786 | 0.01 |
ENSDART00000019997
|
mkrn2
|
makorin, ring finger protein, 2 |
| chr6_+_48862 | 0.01 |
ENSDART00000082954
|
mbd5
|
methyl-CpG binding domain protein 5 |
| chr5_+_457729 | 0.01 |
ENSDART00000025183
|
ift74
|
intraflagellar transport 74 |
| chr22_+_35275206 | 0.01 |
ENSDART00000112234
|
rubcn
|
RUN domain and cysteine-rich domain containing, Beclin 1-interacting protein |
| chr19_-_42045372 | 0.01 |
ENSDART00000144275
|
trioa
|
trio Rho guanine nucleotide exchange factor a |
| chr5_+_393738 | 0.01 |
ENSDART00000161456
|
june
|
JunE proto-oncogene, AP-1 transcription factor subunit |
| chr21_-_45685063 | 0.01 |
ENSDART00000162422
|
sar1b
|
secretion associated, Ras related GTPase 1B |
| chr7_+_28612671 | 0.00 |
ENSDART00000019991
|
slc7a6os
|
solute carrier family 7, member 6 opposite strand |
| chr7_+_65272470 | 0.00 |
ENSDART00000162471
|
bco1
|
beta-carotene oxygenase 1 |
| chr6_+_49771372 | 0.00 |
ENSDART00000063251
|
ctsz
|
cathepsin Z |
| chr17_-_10738001 | 0.00 |
ENSDART00000051526
|
jmjd7
|
jumonji domain containing 7 |
| chr5_+_32009956 | 0.00 |
ENSDART00000188482
|
scai
|
suppressor of cancer cell invasion |
| chr23_-_7052362 | 0.00 |
ENSDART00000127702
ENSDART00000192468 |
prpf6
|
PRP6 pre-mRNA processing factor 6 homolog (S. cerevisiae) |
| chr25_+_5983430 | 0.00 |
ENSDART00000074814
|
ppib
|
peptidylprolyl isomerase B (cyclophilin B) |
| chr22_+_35275468 | 0.00 |
ENSDART00000189516
ENSDART00000181572 ENSDART00000165353 ENSDART00000185352 |
rubcn
|
RUN domain and cysteine-rich domain containing, Beclin 1-interacting protein |
| chr16_-_40373836 | 0.00 |
ENSDART00000134498
|
si:dkey-242e21.3
|
si:dkey-242e21.3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of mbd2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.1 | GO:1903792 | histamine metabolic process(GO:0001692) negative regulation of anion transport(GO:1903792) |
| 0.0 | 0.1 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.0 | 0.1 | GO:0097435 | fibril organization(GO:0097435) |
| 0.0 | 0.0 | GO:0014014 | negative regulation of gliogenesis(GO:0014014) |
| 0.0 | 0.1 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.0 | 0.1 | GO:0061439 | renal system vasculature morphogenesis(GO:0061438) kidney vasculature morphogenesis(GO:0061439) glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.0 | 0.1 | GO:0060546 | negative regulation of necroptotic process(GO:0060546) negative regulation of necrotic cell death(GO:0060547) |
| 0.0 | 0.1 | GO:0006537 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.0 | 0.1 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.0 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) negative regulation of striated muscle contraction(GO:0045988) relaxation of skeletal muscle(GO:0090076) |
| 0.0 | 0.0 | GO:0009912 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.0 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.1 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.0 | 0.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |