Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
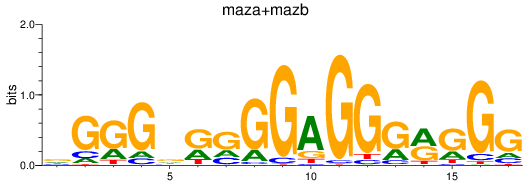
Results for maza+mazb
Z-value: 4.32
Transcription factors associated with maza+mazb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mazb
|
ENSDARG00000063555 | si_ch211-166g5.4 |
|
maza
|
ENSDARG00000087330 | MYC-associated zinc finger protein a (purine-binding transcription factor) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| si:ch211-166g5.4 | dr11_v1_chr12_-_3778848_3778848 | -0.69 | 4.2e-02 | Click! |
| maza | dr11_v1_chr3_-_21118969_21118969 | -0.56 | 1.2e-01 | Click! |
Activity profile of maza+mazb motif
Sorted Z-values of maza+mazb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_44183451 | 8.83 |
ENSDART00000111246
|
cadm3
|
cell adhesion molecule 3 |
| chr2_-_44183613 | 8.77 |
ENSDART00000079596
|
cadm3
|
cell adhesion molecule 3 |
| chr18_-_16123222 | 8.11 |
ENSDART00000061189
|
sspn
|
sarcospan (Kras oncogene-associated gene) |
| chr1_-_39943596 | 7.95 |
ENSDART00000149730
|
stox2a
|
storkhead box 2a |
| chr21_+_26697536 | 6.53 |
ENSDART00000004109
|
gng3
|
guanine nucleotide binding protein (G protein), gamma 3 |
| chr10_-_44026369 | 6.50 |
ENSDART00000185456
|
crybb1
|
crystallin, beta B1 |
| chr19_-_5254699 | 6.11 |
ENSDART00000081951
|
stx1b
|
syntaxin 1B |
| chr14_-_9522364 | 5.75 |
ENSDART00000054689
|
atoh8
|
atonal bHLH transcription factor 8 |
| chr3_-_61162750 | 5.58 |
ENSDART00000055064
|
pvalb8
|
parvalbumin 8 |
| chr7_+_36552725 | 5.25 |
ENSDART00000173682
|
chd9
|
chromodomain helicase DNA binding protein 9 |
| chr5_-_14326959 | 5.05 |
ENSDART00000137355
|
tet3
|
tet methylcytosine dioxygenase 3 |
| chr21_-_43015383 | 5.02 |
ENSDART00000065097
|
dpysl3
|
dihydropyrimidinase-like 3 |
| chr7_-_60831082 | 4.92 |
ENSDART00000073654
ENSDART00000136999 |
pcxb
|
pyruvate carboxylase b |
| chr7_+_39401388 | 4.91 |
ENSDART00000144750
|
tnni2b.1
|
troponin I type 2b (skeletal, fast), tandem duplicate 1 |
| chr25_+_37366698 | 4.86 |
ENSDART00000165400
ENSDART00000192589 |
slc1a2b
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2b |
| chr20_-_40370736 | 4.75 |
ENSDART00000041229
|
fabp7b
|
fatty acid binding protein 7, brain, b |
| chr7_+_44713135 | 4.70 |
ENSDART00000170721
|
si:dkey-56m19.5
|
si:dkey-56m19.5 |
| chr14_-_25577094 | 4.63 |
ENSDART00000163669
|
cplx2
|
complexin 2 |
| chr4_-_16406737 | 4.57 |
ENSDART00000013085
|
dcn
|
decorin |
| chr11_-_11518469 | 4.55 |
ENSDART00000104254
|
krt15
|
keratin 15 |
| chr8_+_28527776 | 4.52 |
ENSDART00000053782
|
scrt2
|
scratch family zinc finger 2 |
| chr11_-_32723851 | 4.50 |
ENSDART00000155592
|
pcdh17
|
protocadherin 17 |
| chr7_-_58098814 | 4.40 |
ENSDART00000147287
ENSDART00000043984 |
ank2b
|
ankyrin 2b, neuronal |
| chr10_+_31244619 | 4.30 |
ENSDART00000145562
ENSDART00000184412 |
robo4
|
roundabout, axon guidance receptor, homolog 4 (Drosophila) |
| chr14_-_39074539 | 4.26 |
ENSDART00000030509
|
glra4a
|
glycine receptor, alpha 4a |
| chr12_-_16764751 | 4.24 |
ENSDART00000113862
|
zgc:174154
|
zgc:174154 |
| chr3_-_46811611 | 4.23 |
ENSDART00000134092
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr17_+_26569601 | 4.21 |
ENSDART00000153897
|
ndnfl
|
neuron-derived neurotrophic factor , like |
| chr20_+_15015557 | 4.16 |
ENSDART00000039345
|
myoc
|
myocilin |
| chr1_-_38709551 | 4.15 |
ENSDART00000128794
|
gpm6ab
|
glycoprotein M6Ab |
| chr14_-_1990290 | 4.14 |
ENSDART00000183382
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr3_+_27027781 | 4.07 |
ENSDART00000065495
|
emp2
|
epithelial membrane protein 2 |
| chr8_+_6954984 | 4.02 |
ENSDART00000145610
|
si:ch211-255g12.6
|
si:ch211-255g12.6 |
| chr17_-_44247707 | 3.96 |
ENSDART00000126097
|
otx2b
|
orthodenticle homeobox 2b |
| chr23_-_35756115 | 3.95 |
ENSDART00000043429
|
jph2
|
junctophilin 2 |
| chr4_-_12862087 | 3.93 |
ENSDART00000080536
|
hmga2
|
high mobility group AT-hook 2 |
| chr16_+_23431189 | 3.93 |
ENSDART00000004679
|
icn
|
ictacalcin |
| chr10_+_22724059 | 3.91 |
ENSDART00000136123
|
kdm6bb
|
lysine (K)-specific demethylase 6B, b |
| chr12_+_32388860 | 3.69 |
ENSDART00000075568
|
nog3
|
noggin 3 |
| chr6_-_20875111 | 3.65 |
ENSDART00000115118
ENSDART00000159916 |
tns1a
|
tensin 1a |
| chr5_+_9348284 | 3.64 |
ENSDART00000149417
|
tal2
|
T-cell acute lymphocytic leukemia 2 |
| chr18_-_45736 | 3.63 |
ENSDART00000148373
ENSDART00000148950 |
gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr11_+_36989696 | 3.63 |
ENSDART00000045888
|
tkta
|
transketolase a |
| chr5_-_14344647 | 3.57 |
ENSDART00000188456
|
tet3
|
tet methylcytosine dioxygenase 3 |
| chr14_+_34486629 | 3.55 |
ENSDART00000131861
|
tmsb2
|
thymosin beta 2 |
| chr1_+_37391141 | 3.52 |
ENSDART00000083593
ENSDART00000168647 |
sparcl1
|
SPARC-like 1 |
| chr13_-_31435137 | 3.44 |
ENSDART00000057441
|
rtn1a
|
reticulon 1a |
| chr1_+_45351890 | 3.44 |
ENSDART00000145486
|
si:ch211-243a20.3
|
si:ch211-243a20.3 |
| chr18_-_46010 | 3.42 |
ENSDART00000052641
|
gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr18_-_44610992 | 3.39 |
ENSDART00000125968
ENSDART00000185836 |
spred3
|
sprouty-related, EVH1 domain containing 3 |
| chr19_+_3001473 | 3.39 |
ENSDART00000145363
|
scxa
|
scleraxis bHLH transcription factor a |
| chr17_+_52822422 | 3.37 |
ENSDART00000158273
ENSDART00000161414 |
meis2a
|
Meis homeobox 2a |
| chr13_+_23214100 | 3.34 |
ENSDART00000163393
|
sorbs1
|
sorbin and SH3 domain containing 1 |
| chr24_-_4973765 | 3.32 |
ENSDART00000127597
|
zic1
|
zic family member 1 (odd-paired homolog, Drosophila) |
| chr18_-_39583601 | 3.32 |
ENSDART00000125116
|
tnfaip8l3
|
tumor necrosis factor, alpha-induced protein 8-like 3 |
| chr8_+_46217861 | 3.26 |
ENSDART00000038790
|
angptl7
|
angiopoietin-like 7 |
| chr3_-_38692920 | 3.23 |
ENSDART00000155042
|
mpp3a
|
membrane protein, palmitoylated 3a (MAGUK p55 subfamily member 3) |
| chr19_-_1961024 | 3.22 |
ENSDART00000108784
|
mturn
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr10_-_27197044 | 3.22 |
ENSDART00000137928
|
auts2a
|
autism susceptibility candidate 2a |
| chr13_+_41022502 | 3.18 |
ENSDART00000026808
|
dkk1a
|
dickkopf WNT signaling pathway inhibitor 1a |
| chr3_+_23687909 | 3.17 |
ENSDART00000046638
|
hoxb8a
|
homeobox B8a |
| chr16_-_36798783 | 3.15 |
ENSDART00000145697
|
calb1
|
calbindin 1 |
| chr3_-_28075756 | 3.14 |
ENSDART00000122037
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr22_+_38159823 | 3.08 |
ENSDART00000104527
|
tm4sf18
|
transmembrane 4 L six family member 18 |
| chr19_+_4912817 | 3.07 |
ENSDART00000101658
ENSDART00000165082 |
ppp1r1b
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr11_-_97817 | 3.04 |
ENSDART00000092903
|
elmo2
|
engulfment and cell motility 2 |
| chr12_-_31009315 | 3.03 |
ENSDART00000133854
|
tcf7l2
|
transcription factor 7 like 2 |
| chr2_-_9646857 | 3.01 |
ENSDART00000056901
|
zgc:153615
|
zgc:153615 |
| chr23_+_21459263 | 2.97 |
ENSDART00000104209
|
her4.3
|
hairy-related 4, tandem duplicate 3 |
| chr8_+_22931427 | 2.95 |
ENSDART00000063096
|
sypa
|
synaptophysin a |
| chr5_+_38263240 | 2.92 |
ENSDART00000051231
|
gnb2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr15_+_47903864 | 2.91 |
ENSDART00000063835
|
otx5
|
orthodenticle homolog 5 |
| chr3_+_23726148 | 2.91 |
ENSDART00000174580
|
hoxb3a
|
homeobox B3a |
| chr4_+_2619132 | 2.89 |
ENSDART00000128807
|
gpr22a
|
G protein-coupled receptor 22a |
| chr14_-_30642819 | 2.88 |
ENSDART00000078154
|
npas4a
|
neuronal PAS domain protein 4a |
| chr12_-_16941319 | 2.86 |
ENSDART00000109968
|
zgc:174855
|
zgc:174855 |
| chr3_-_39152478 | 2.83 |
ENSDART00000154550
|
si:dkeyp-57f11.2
|
si:dkeyp-57f11.2 |
| chr19_-_2231146 | 2.83 |
ENSDART00000181909
ENSDART00000043595 |
twist1a
|
twist family bHLH transcription factor 1a |
| chr18_+_44649804 | 2.81 |
ENSDART00000059063
|
ehd2b
|
EH-domain containing 2b |
| chr19_+_30633453 | 2.79 |
ENSDART00000052124
|
fam49al
|
family with sequence similarity 49, member A-like |
| chr9_+_13999620 | 2.75 |
ENSDART00000143229
|
cd28l
|
cd28-like molecule |
| chr6_-_11780070 | 2.75 |
ENSDART00000151195
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr25_-_11088839 | 2.75 |
ENSDART00000154748
|
sv2bb
|
synaptic vesicle glycoprotein 2Bb |
| chr25_+_1732838 | 2.75 |
ENSDART00000159555
ENSDART00000168161 |
FBLN1
|
fibulin 1 |
| chr14_+_34547554 | 2.74 |
ENSDART00000074819
|
gabrp
|
gamma-aminobutyric acid (GABA) A receptor, pi |
| chr6_+_22597362 | 2.70 |
ENSDART00000131242
|
cygb2
|
cytoglobin 2 |
| chr5_-_28625515 | 2.70 |
ENSDART00000190782
ENSDART00000179736 ENSDART00000131729 |
tnc
|
tenascin C |
| chr12_-_26851726 | 2.70 |
ENSDART00000047724
|
zeb1b
|
zinc finger E-box binding homeobox 1b |
| chr12_-_10705916 | 2.70 |
ENSDART00000164038
|
FO704786.1
|
|
| chr1_-_25936677 | 2.68 |
ENSDART00000146488
ENSDART00000136321 |
myoz2b
|
myozenin 2b |
| chr18_-_50152689 | 2.68 |
ENSDART00000006078
|
loxl1
|
lysyl oxidase-like 1 |
| chr21_+_5169154 | 2.67 |
ENSDART00000102559
|
zgc:122979
|
zgc:122979 |
| chr7_+_26029672 | 2.65 |
ENSDART00000101126
|
alox12
|
arachidonate 12-lipoxygenase |
| chr15_+_1397811 | 2.65 |
ENSDART00000102125
|
schip1
|
schwannomin interacting protein 1 |
| chr10_-_22803740 | 2.64 |
ENSDART00000079469
ENSDART00000187968 ENSDART00000122543 |
pcolcea
|
procollagen C-endopeptidase enhancer a |
| chr4_-_17055782 | 2.64 |
ENSDART00000134595
|
sox5
|
SRY (sex determining region Y)-box 5 |
| chr24_+_41931585 | 2.63 |
ENSDART00000130310
|
epb41l3a
|
erythrocyte membrane protein band 4.1-like 3a |
| chr16_-_23800484 | 2.62 |
ENSDART00000139964
|
rps27.2
|
ribosomal protein S27, isoform 2 |
| chr4_+_12612145 | 2.62 |
ENSDART00000181201
|
lmo3
|
LIM domain only 3 |
| chr10_-_35542071 | 2.62 |
ENSDART00000162139
|
si:ch211-244c8.4
|
si:ch211-244c8.4 |
| chr18_+_21122818 | 2.59 |
ENSDART00000060015
ENSDART00000060184 |
chka
|
choline kinase alpha |
| chr13_-_45523026 | 2.59 |
ENSDART00000020663
|
rhd
|
Rh blood group, D antigen |
| chr23_-_31428763 | 2.52 |
ENSDART00000053545
|
zgc:153284
|
zgc:153284 |
| chr10_+_10210455 | 2.52 |
ENSDART00000144214
|
sh2d3ca
|
SH2 domain containing 3Ca |
| chr9_+_14152211 | 2.52 |
ENSDART00000148055
|
si:ch211-67e16.11
|
si:ch211-67e16.11 |
| chr15_-_2188332 | 2.51 |
ENSDART00000138941
ENSDART00000009564 |
shox2
|
short stature homeobox 2 |
| chr20_+_15982482 | 2.51 |
ENSDART00000020999
|
angptl1a
|
angiopoietin-like 1a |
| chr25_-_12788370 | 2.49 |
ENSDART00000158551
|
slc7a5
|
solute carrier family 7 (amino acid transporter light chain, L system), member 5 |
| chr14_+_24283915 | 2.47 |
ENSDART00000172868
|
klhl3
|
kelch-like family member 3 |
| chr13_+_28854438 | 2.46 |
ENSDART00000193407
ENSDART00000189554 |
CU639469.1
|
|
| chr11_-_3334248 | 2.45 |
ENSDART00000154314
ENSDART00000121861 |
prph
|
peripherin |
| chr2_+_22694382 | 2.45 |
ENSDART00000139196
|
kif1ab
|
kinesin family member 1Ab |
| chr20_+_20672163 | 2.41 |
ENSDART00000027758
|
rtn1b
|
reticulon 1b |
| chr4_-_17055950 | 2.39 |
ENSDART00000162945
|
sox5
|
SRY (sex determining region Y)-box 5 |
| chr11_-_44543082 | 2.39 |
ENSDART00000099568
|
gpr137bb
|
G protein-coupled receptor 137Bb |
| chr3_+_60721342 | 2.39 |
ENSDART00000157772
|
foxj1a
|
forkhead box J1a |
| chr7_+_46261199 | 2.38 |
ENSDART00000170390
ENSDART00000183227 |
znf536
|
zinc finger protein 536 |
| chr3_-_22212764 | 2.37 |
ENSDART00000155490
|
maptb
|
microtubule-associated protein tau b |
| chr1_-_29045426 | 2.36 |
ENSDART00000019770
|
gpm6ba
|
glycoprotein M6Ba |
| chr15_+_47440477 | 2.35 |
ENSDART00000002384
|
phox2a
|
paired-like homeobox 2a |
| chr9_-_14108896 | 2.35 |
ENSDART00000135209
|
prkag3b
|
protein kinase, AMP-activated, gamma 3b non-catalytic subunit |
| chr1_-_22861348 | 2.35 |
ENSDART00000139412
|
SMIM18
|
si:dkey-92j12.6 |
| chr11_-_21304452 | 2.34 |
ENSDART00000163008
|
RASSF5
|
si:dkey-85p17.3 |
| chr16_+_7662609 | 2.29 |
ENSDART00000184895
ENSDART00000149404 ENSDART00000081418 ENSDART00000081422 |
bves
|
blood vessel epicardial substance |
| chr3_-_28120092 | 2.28 |
ENSDART00000151143
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr16_-_21785261 | 2.28 |
ENSDART00000078858
|
si:ch73-86n18.1
|
si:ch73-86n18.1 |
| chr4_+_20255160 | 2.27 |
ENSDART00000188658
|
lrtm2a
|
leucine-rich repeats and transmembrane domains 2a |
| chr14_-_2602445 | 2.26 |
ENSDART00000166910
|
etf1a
|
eukaryotic translation termination factor 1a |
| chr25_+_31323978 | 2.24 |
ENSDART00000067030
|
lsp1
|
lymphocyte-specific protein 1 |
| chr16_-_26296477 | 2.23 |
ENSDART00000157553
|
erfl1
|
Ets2 repressor factor like 1 |
| chr16_+_24842662 | 2.22 |
ENSDART00000157333
|
si:dkey-79d12.6
|
si:dkey-79d12.6 |
| chr7_+_44445595 | 2.21 |
ENSDART00000108766
|
cdh5
|
cadherin 5 |
| chr15_-_3736773 | 2.20 |
ENSDART00000090624
|
lpar6a
|
lysophosphatidic acid receptor 6a |
| chr13_+_22480496 | 2.20 |
ENSDART00000136863
ENSDART00000131870 ENSDART00000078720 ENSDART00000078740 ENSDART00000139218 |
ldb3a
|
LIM domain binding 3a |
| chr4_+_19534833 | 2.18 |
ENSDART00000140028
|
lrrc4.1
|
leucine rich repeat containing 4.1 |
| chr2_-_32555625 | 2.16 |
ENSDART00000056641
ENSDART00000137531 |
smarcd3a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3a |
| chr22_+_10090673 | 2.15 |
ENSDART00000186680
|
si:dkey-102c8.3
|
si:dkey-102c8.3 |
| chr3_+_16265924 | 2.15 |
ENSDART00000122519
|
st8sia6
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 6 |
| chr14_+_30730749 | 2.15 |
ENSDART00000087884
|
ccdc85b
|
coiled-coil domain containing 85B |
| chr15_-_23342752 | 2.14 |
ENSDART00000020425
|
mcamb
|
melanoma cell adhesion molecule b |
| chr1_+_44127292 | 2.14 |
ENSDART00000160542
|
cabp2a
|
calcium binding protein 2a |
| chr3_-_28250722 | 2.12 |
ENSDART00000165936
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr4_-_6459863 | 2.11 |
ENSDART00000138367
|
foxp2
|
forkhead box P2 |
| chr3_-_30625219 | 2.10 |
ENSDART00000151698
|
syt3
|
synaptotagmin III |
| chr21_-_28901095 | 2.10 |
ENSDART00000180820
|
cxxc5a
|
CXXC finger protein 5a |
| chr11_-_11471857 | 2.09 |
ENSDART00000030103
|
krt94
|
keratin 94 |
| chr2_+_43469241 | 2.07 |
ENSDART00000142078
ENSDART00000098265 |
nrp1b
|
neuropilin 1b |
| chr2_-_43168292 | 2.07 |
ENSDART00000132588
|
crema
|
cAMP responsive element modulator a |
| chr4_-_1360495 | 2.06 |
ENSDART00000164623
|
ptn
|
pleiotrophin |
| chr21_-_21373242 | 2.05 |
ENSDART00000079629
|
ppm1nb
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Nb (putative) |
| chr5_-_67878064 | 2.02 |
ENSDART00000111203
|
tagln3a
|
transgelin 3a |
| chr21_+_19648814 | 2.01 |
ENSDART00000048581
|
fgf10a
|
fibroblast growth factor 10a |
| chr19_-_26863626 | 2.00 |
ENSDART00000145568
|
prrt1
|
proline-rich transmembrane protein 1 |
| chr19_+_9533008 | 1.99 |
ENSDART00000104607
|
fam131ba
|
family with sequence similarity 131, member Ba |
| chr2_-_56095275 | 1.98 |
ENSDART00000154701
|
si:ch211-178n15.1
|
si:ch211-178n15.1 |
| chr21_-_9991502 | 1.97 |
ENSDART00000169320
|
esm1
|
endothelial cell-specific molecule 1 |
| chr6_+_55365632 | 1.97 |
ENSDART00000169240
|
pltp
|
phospholipid transfer protein |
| chr23_+_14217508 | 1.96 |
ENSDART00000143618
|
birc7
|
baculoviral IAP repeat containing 7 |
| chr20_+_37661229 | 1.95 |
ENSDART00000138539
|
aig1
|
androgen-induced 1 (H. sapiens) |
| chr19_-_5125663 | 1.95 |
ENSDART00000005547
ENSDART00000132642 |
gnb3b
|
guanine nucleotide binding protein (G protein), beta polypeptide 3b |
| chr16_-_16590780 | 1.95 |
ENSDART00000059841
|
si:ch211-257p13.3
|
si:ch211-257p13.3 |
| chr15_+_19838458 | 1.95 |
ENSDART00000101204
|
alcamb
|
activated leukocyte cell adhesion molecule b |
| chr7_+_19495379 | 1.93 |
ENSDART00000180514
|
si:ch211-212k18.8
|
si:ch211-212k18.8 |
| chr19_-_27777228 | 1.93 |
ENSDART00000046166
|
adcy2b
|
adenylate cyclase 2b (brain) |
| chr18_+_6054816 | 1.91 |
ENSDART00000113668
|
si:ch73-386h18.1
|
si:ch73-386h18.1 |
| chr2_-_50372467 | 1.90 |
ENSDART00000108900
|
cntnap2b
|
contactin associated protein like 2b |
| chr4_+_11311315 | 1.89 |
ENSDART00000051792
|
sema3aa
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Aa |
| chr1_+_51721851 | 1.89 |
ENSDART00000040397
|
prdx2
|
peroxiredoxin 2 |
| chr12_-_34459601 | 1.89 |
ENSDART00000111717
|
fscn2b
|
fascin actin-bundling protein 2b, retinal |
| chr25_+_25464630 | 1.88 |
ENSDART00000150537
|
rassf7a
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7a |
| chr20_+_37661547 | 1.87 |
ENSDART00000007253
|
aig1
|
androgen-induced 1 (H. sapiens) |
| chr10_-_26729930 | 1.87 |
ENSDART00000145532
|
fgf13b
|
fibroblast growth factor 13b |
| chr9_+_21407987 | 1.86 |
ENSDART00000145627
|
gja3
|
gap junction protein, alpha 3 |
| chr20_+_33875256 | 1.86 |
ENSDART00000002554
|
rxrgb
|
retinoid X receptor, gamma b |
| chr21_+_45841731 | 1.85 |
ENSDART00000038657
|
faxdc2
|
fatty acid hydroxylase domain containing 2 |
| chr6_-_39275793 | 1.85 |
ENSDART00000180477
ENSDART00000148531 |
arhgef25b
|
Rho guanine nucleotide exchange factor (GEF) 25b |
| chr17_-_29902187 | 1.83 |
ENSDART00000009104
|
esrrb
|
estrogen-related receptor beta |
| chr7_-_41468942 | 1.81 |
ENSDART00000174087
|
smarcd3b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3b |
| chr19_-_41213718 | 1.80 |
ENSDART00000077121
|
pdk4
|
pyruvate dehydrogenase kinase, isozyme 4 |
| chr15_-_33527631 | 1.80 |
ENSDART00000187646
ENSDART00000161151 |
stard13b
|
StAR-related lipid transfer (START) domain containing 13b |
| chr4_+_14360372 | 1.80 |
ENSDART00000007103
|
nuak1a
|
NUAK family, SNF1-like kinase, 1a |
| chr3_-_36115339 | 1.79 |
ENSDART00000187406
ENSDART00000123505 ENSDART00000151775 |
rab11fip4a
|
RAB11 family interacting protein 4 (class II) a |
| chr13_+_22480857 | 1.79 |
ENSDART00000078721
ENSDART00000044719 ENSDART00000130957 ENSDART00000078757 ENSDART00000130424 ENSDART00000078747 |
ldb3a
|
LIM domain binding 3a |
| chr11_-_25829712 | 1.78 |
ENSDART00000103549
|
skib
|
v-ski avian sarcoma viral oncogene homolog b |
| chr21_-_43117327 | 1.78 |
ENSDART00000122352
|
p4ha2
|
procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha polypeptide 2 |
| chr5_-_5831037 | 1.76 |
ENSDART00000112856
|
zmp:0000000846
|
zmp:0000000846 |
| chr14_+_42172257 | 1.76 |
ENSDART00000074362
|
pcdh18b
|
protocadherin 18b |
| chr12_-_34887943 | 1.73 |
ENSDART00000027379
|
bicral
|
BRD4 interacting chromatin remodeling complex associated protein like |
| chr3_-_19899914 | 1.71 |
ENSDART00000134969
|
rnd2
|
Rho family GTPase 2 |
| chr3_+_32553714 | 1.71 |
ENSDART00000165638
|
pax10
|
paired box 10 |
| chr19_+_16032383 | 1.71 |
ENSDART00000046530
|
rab42a
|
RAB42, member RAS oncogene family a |
| chr18_+_49225552 | 1.71 |
ENSDART00000135026
|
si:ch211-136a13.1
|
si:ch211-136a13.1 |
| chr17_-_46457622 | 1.70 |
ENSDART00000130215
|
TMEM179 (1 of many)
|
transmembrane protein 179 |
| chr4_-_9173552 | 1.70 |
ENSDART00000042963
|
chst11
|
carbohydrate (chondroitin 4) sulfotransferase 11 |
| chr18_+_12058403 | 1.69 |
ENSDART00000140854
ENSDART00000193632 ENSDART00000190519 ENSDART00000190685 ENSDART00000112671 |
bicd1a
|
bicaudal D homolog 1a |
| chr3_-_33417826 | 1.68 |
ENSDART00000084284
|
abi3a
|
ABI family, member 3a |
| chr10_+_34394454 | 1.67 |
ENSDART00000110121
|
stard13a
|
StAR-related lipid transfer (START) domain containing 13a |
| chr17_-_26926577 | 1.67 |
ENSDART00000050202
|
rcan3
|
regulator of calcineurin 3 |
| chr13_-_50139916 | 1.66 |
ENSDART00000099475
|
nid1a
|
nidogen 1a |
| chr3_-_30685401 | 1.65 |
ENSDART00000151097
|
si:ch211-51c14.1
|
si:ch211-51c14.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of maza+mazb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.1 | GO:0006601 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 1.9 | 5.7 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 1.7 | 8.6 | GO:0032616 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 1.4 | 4.3 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 1.4 | 4.1 | GO:0003093 | renal system process involved in regulation of blood volume(GO:0001977) renal system process involved in regulation of systemic arterial blood pressure(GO:0003071) regulation of glomerular filtration(GO:0003093) |
| 1.2 | 6.1 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 1.1 | 3.4 | GO:0035992 | tendon formation(GO:0035992) |
| 0.9 | 5.7 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.9 | 5.6 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.9 | 2.7 | GO:0014814 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) |
| 0.9 | 4.4 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.8 | 2.5 | GO:1903792 | regulation of neurotransmitter uptake(GO:0051580) negative regulation of anion transport(GO:1903792) |
| 0.8 | 2.5 | GO:0072156 | distal tubule morphogenesis(GO:0072156) |
| 0.8 | 2.4 | GO:0036378 | calcitriol biosynthetic process from calciol(GO:0036378) vitamin D biosynthetic process(GO:0042368) cellular alcohol metabolic process(GO:0044107) cellular alcohol biosynthetic process(GO:0044108) |
| 0.8 | 2.4 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.8 | 3.2 | GO:1900271 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 0.8 | 3.0 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.7 | 3.0 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.7 | 3.6 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.7 | 2.1 | GO:0070857 | regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.7 | 2.0 | GO:0034375 | high-density lipoprotein particle remodeling(GO:0034375) |
| 0.6 | 3.2 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.6 | 2.5 | GO:0060437 | lung growth(GO:0060437) secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.6 | 1.9 | GO:0048569 | post-embryonic organ development(GO:0048569) |
| 0.6 | 5.4 | GO:0032986 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.6 | 2.9 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) |
| 0.6 | 3.4 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.6 | 4.5 | GO:0043516 | regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043516) |
| 0.6 | 2.2 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.5 | 2.7 | GO:0070587 | regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 0.5 | 2.1 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.5 | 6.2 | GO:0050936 | xanthophore differentiation(GO:0050936) |
| 0.5 | 2.4 | GO:0060295 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.5 | 3.3 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 0.5 | 2.3 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.4 | 1.3 | GO:0048682 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.4 | 2.6 | GO:0003272 | endocardial cushion morphogenesis(GO:0003203) endocardial cushion formation(GO:0003272) |
| 0.4 | 5.0 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.4 | 3.9 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.4 | 4.3 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.4 | 1.9 | GO:0099612 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.4 | 1.9 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.4 | 4.1 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.4 | 3.6 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.3 | 2.6 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.3 | 1.0 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.3 | 4.5 | GO:0001952 | regulation of cell-matrix adhesion(GO:0001952) |
| 0.3 | 0.9 | GO:0014014 | negative regulation of gliogenesis(GO:0014014) |
| 0.3 | 2.9 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.3 | 2.6 | GO:1904861 | excitatory synapse assembly(GO:1904861) |
| 0.3 | 1.4 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.3 | 1.7 | GO:0060632 | regulation of microtubule-based movement(GO:0060632) |
| 0.3 | 1.9 | GO:0021885 | forebrain cell migration(GO:0021885) |
| 0.3 | 1.4 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.3 | 0.8 | GO:0021531 | spinal cord radial glial cell differentiation(GO:0021531) eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.3 | 4.9 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.3 | 1.3 | GO:0050868 | negative regulation of T cell activation(GO:0050868) negative regulation of leukocyte cell-cell adhesion(GO:1903038) |
| 0.3 | 1.0 | GO:2000742 | cytidine to uridine editing(GO:0016554) regulation of anterior head development(GO:2000742) |
| 0.2 | 1.2 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.2 | 1.0 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.2 | 1.2 | GO:0006691 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.2 | 4.0 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.2 | 3.7 | GO:0035844 | cloaca development(GO:0035844) |
| 0.2 | 1.4 | GO:0010752 | regulation of cGMP-mediated signaling(GO:0010752) |
| 0.2 | 2.6 | GO:0051121 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.2 | 2.3 | GO:0021703 | locus ceruleus development(GO:0021703) |
| 0.2 | 4.6 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.2 | 1.9 | GO:0000303 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.2 | 1.7 | GO:0070571 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.2 | 2.8 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.2 | 6.3 | GO:0050654 | chondroitin sulfate proteoglycan metabolic process(GO:0050654) |
| 0.2 | 3.2 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.2 | 7.5 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.2 | 0.9 | GO:0050996 | positive regulation of lipid catabolic process(GO:0050996) |
| 0.2 | 3.2 | GO:1990399 | sensory epithelium regeneration(GO:0070654) epithelium regeneration(GO:1990399) |
| 0.2 | 0.7 | GO:0055016 | hypochord development(GO:0055016) |
| 0.2 | 3.0 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.2 | 2.1 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.2 | 2.7 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.2 | 1.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.2 | 1.4 | GO:0016576 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.1 | 0.6 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.1 | 2.5 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 2.7 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 1.9 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 3.0 | GO:0071910 | determination of liver left/right asymmetry(GO:0071910) |
| 0.1 | 3.6 | GO:0021514 | ventral spinal cord interneuron differentiation(GO:0021514) |
| 0.1 | 1.5 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 | 3.9 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.1 | 1.2 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 | 4.5 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.1 | 2.5 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.1 | 2.6 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.8 | GO:0070293 | renal absorption(GO:0070293) |
| 0.1 | 0.5 | GO:0031649 | heat generation(GO:0031649) regulation of heat generation(GO:0031650) positive regulation of heat generation(GO:0031652) regulation of phospholipid biosynthetic process(GO:0071071) negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.1 | 4.6 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 1.1 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.1 | 2.3 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.1 | 0.4 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.1 | 3.8 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.1 | 4.9 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 0.9 | GO:0010002 | cardioblast differentiation(GO:0010002) negative regulation of muscle organ development(GO:0048635) |
| 0.1 | 4.0 | GO:0051588 | regulation of neurotransmitter transport(GO:0051588) |
| 0.1 | 0.8 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.1 | 1.5 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.1 | 1.1 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.1 | 0.6 | GO:0021627 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.1 | 0.9 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 1.9 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.1 | 4.8 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 2.3 | GO:0031103 | axon regeneration(GO:0031103) |
| 0.1 | 2.6 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.1 | 0.7 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 1.6 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.1 | 0.7 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.1 | 0.8 | GO:0097369 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 4.8 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.1 | 1.1 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.1 | 2.8 | GO:0045103 | intermediate filament-based process(GO:0045103) intermediate filament cytoskeleton organization(GO:0045104) |
| 0.1 | 0.9 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.8 | GO:0030520 | intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.1 | 0.7 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 3.7 | GO:0042129 | regulation of T cell proliferation(GO:0042129) |
| 0.1 | 2.8 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.1 | 0.8 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.1 | 1.3 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 3.8 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.1 | 2.1 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.1 | 1.8 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.1 | 4.6 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 1.5 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.1 | 1.6 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 0.5 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.1 | 0.9 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 1.4 | GO:0008214 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.1 | 2.0 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.1 | 0.2 | GO:1901017 | negative regulation of potassium ion transmembrane transporter activity(GO:1901017) negative regulation of potassium ion transmembrane transport(GO:1901380) negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.1 | 0.5 | GO:0048922 | posterior lateral line neuromast deposition(GO:0048922) |
| 0.1 | 0.3 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 6.5 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.1 | GO:0051610 | serotonin transport(GO:0006837) serotonin uptake(GO:0051610) |
| 0.0 | 0.1 | GO:2000108 | regulation of platelet activation(GO:0010543) regulation of platelet aggregation(GO:0090330) positive regulation of leukocyte apoptotic process(GO:2000108) |
| 0.0 | 0.3 | GO:1902269 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.6 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.4 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 1.5 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.9 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 1.3 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.0 | 0.9 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 1.9 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.0 | 3.2 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 1.1 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 1.8 | GO:0010906 | regulation of glucose metabolic process(GO:0010906) |
| 0.0 | 1.7 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.9 | GO:0071222 | cellular response to molecule of bacterial origin(GO:0071219) cellular response to lipopolysaccharide(GO:0071222) |
| 0.0 | 7.6 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.1 | GO:0039020 | pronephric nephron tubule development(GO:0039020) |
| 0.0 | 0.5 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 4.8 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.0 | 2.1 | GO:0051781 | positive regulation of cell division(GO:0051781) |
| 0.0 | 1.8 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 2.3 | GO:0009880 | embryonic pattern specification(GO:0009880) |
| 0.0 | 0.2 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 1.2 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 1.6 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.0 | 2.9 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.3 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.0 | 0.3 | GO:0060078 | regulation of postsynaptic membrane potential(GO:0060078) excitatory postsynaptic potential(GO:0060079) chemical synaptic transmission, postsynaptic(GO:0099565) |
| 0.0 | 1.1 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.3 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.0 | 3.1 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.0 | 3.3 | GO:0007626 | locomotory behavior(GO:0007626) |
| 0.0 | 1.4 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 1.3 | GO:0006909 | phagocytosis(GO:0006909) |
| 0.0 | 0.4 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.7 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 0.1 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.0 | 1.2 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 1.8 | GO:0008016 | regulation of heart contraction(GO:0008016) |
| 0.0 | 5.7 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 0.1 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.0 | 0.2 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 1.7 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.4 | GO:0060117 | auditory receptor cell development(GO:0060117) |
| 0.0 | 0.5 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.0 | 0.7 | GO:0006171 | cAMP biosynthetic process(GO:0006171) |
| 0.0 | 0.4 | GO:0009948 | anterior/posterior axis specification(GO:0009948) |
| 0.0 | 1.7 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 0.9 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.1 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.8 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.9 | GO:0014065 | phosphatidylinositol 3-kinase signaling(GO:0014065) |
| 0.0 | 0.1 | GO:0031102 | neuron projection regeneration(GO:0031102) |
| 0.0 | 2.4 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 0.3 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 1.3 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.7 | GO:0051017 | actin filament bundle assembly(GO:0051017) |
| 0.0 | 0.6 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 0.5 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 0.2 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.2 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.0 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.8 | 3.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.5 | 2.3 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.4 | 6.6 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.4 | 6.5 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.4 | 8.1 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.3 | 1.4 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.3 | 4.1 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.3 | 7.2 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.3 | 4.4 | GO:0030315 | T-tubule(GO:0030315) |
| 0.3 | 6.5 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.3 | 3.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.2 | 1.2 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.2 | 1.9 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.2 | 1.4 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.2 | 3.9 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.2 | 2.6 | GO:0005922 | connexon complex(GO:0005922) |
| 0.2 | 1.5 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.2 | 2.6 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.2 | 5.4 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 2.0 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 1.9 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 4.3 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 4.6 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 4.2 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 3.4 | GO:0098831 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.1 | 4.9 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.9 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 3.0 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 1.3 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.1 | 0.8 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 9.8 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 1.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 4.0 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 1.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 3.7 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 0.9 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 4.8 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 1.0 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 5.6 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 2.9 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.1 | 3.0 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 2.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 3.3 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 1.1 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 11.8 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 3.1 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 1.8 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.4 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 1.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 9.0 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 1.9 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.9 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 47.9 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 0.7 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 1.3 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.0 | 0.7 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.8 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.8 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 5.5 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 2.8 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.7 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.9 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.3 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 3.6 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 1.1 | GO:0070382 | exocytic vesicle(GO:0070382) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.9 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 1.2 | 8.6 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 1.1 | 3.2 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.8 | 2.4 | GO:0031073 | vitamin D3 25-hydroxylase activity(GO:0030343) cholesterol 26-hydroxylase activity(GO:0031073) |
| 0.8 | 2.3 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.7 | 3.0 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.6 | 3.2 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.6 | 3.6 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.5 | 2.0 | GO:1904121 | phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.5 | 4.3 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.4 | 2.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.4 | 5.0 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.4 | 3.9 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.4 | 2.7 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.3 | 1.7 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.3 | 4.7 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.3 | 0.9 | GO:0005131 | growth hormone receptor binding(GO:0005131) growth hormone activity(GO:0070186) |
| 0.3 | 6.5 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.3 | 1.2 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.3 | 4.8 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.3 | 1.2 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.3 | 2.6 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.3 | 1.7 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.3 | 1.9 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.3 | 2.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.3 | 2.7 | GO:0051373 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.3 | 1.9 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.3 | 8.6 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.2 | 4.4 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.2 | 1.8 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.2 | 0.9 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.2 | 1.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.2 | 0.9 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.2 | 7.1 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.2 | 8.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.2 | 1.7 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.2 | 2.5 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.2 | 1.9 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.2 | 4.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.2 | 1.2 | GO:0036374 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.2 | 1.0 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.2 | 7.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.2 | 1.1 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.2 | 2.8 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.2 | 0.9 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.2 | 0.7 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.2 | 5.0 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.2 | 3.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 1.6 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.2 | 2.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.2 | 2.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.2 | 5.4 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.2 | 3.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.2 | 2.7 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.2 | 2.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.2 | 2.0 | GO:0043028 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.1 | 2.1 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 1.4 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 0.3 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 2.3 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 0.8 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.1 | 1.0 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 0.5 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 6.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 3.2 | GO:0016208 | AMP binding(GO:0016208) |
| 0.1 | 1.0 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 2.7 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.1 | 1.0 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 0.5 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.1 | 3.9 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 1.9 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 4.0 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 0.5 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.1 | 4.9 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 1.8 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.1 | 2.5 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 6.3 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 1.0 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.1 | 6.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.5 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 4.2 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 1.9 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 0.8 | GO:0022821 | potassium ion antiporter activity(GO:0022821) |
| 0.1 | 0.2 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.1 | 1.0 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 3.3 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 2.0 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.2 | GO:0042165 | neurotransmitter binding(GO:0042165) acetylcholine binding(GO:0042166) |
| 0.1 | 1.8 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 4.6 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.1 | 1.0 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.1 | 1.1 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 1.5 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 1.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:2001070 | starch binding(GO:2001070) |
| 0.0 | 0.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.4 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 1.2 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.3 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 1.5 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.5 | GO:0030955 | potassium ion binding(GO:0030955) alkali metal ion binding(GO:0031420) |
| 0.0 | 0.2 | GO:0004945 | angiotensin type I receptor activity(GO:0001596) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.7 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.5 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.9 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 1.0 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 1.4 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.9 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 1.3 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 3.7 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 5.2 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.2 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.0 | 0.9 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.6 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.9 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.7 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.5 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.8 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.4 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 25.8 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.9 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 1.0 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.0 | 1.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.8 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 2.0 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 1.6 | GO:0051427 | hormone receptor binding(GO:0051427) |
| 0.0 | 0.5 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 1.1 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 11.5 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.3 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 10.7 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 1.4 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 3.2 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.8 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.1 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.3 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 24.4 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.3 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.8 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 1.9 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.1 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 1.9 | GO:0042277 | peptide binding(GO:0042277) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 2.2 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 2.7 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 1.0 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 1.6 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 1.1 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 0.5 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 1.0 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 3.1 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.1 | 2.7 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 2.6 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 1.0 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 1.0 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 0.7 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 4.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 0.9 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 7.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.1 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 6.3 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.0 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.6 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.1 | PID S1P S1P2 PATHWAY | S1P2 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 9.5 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.5 | 5.0 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.4 | 5.5 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.4 | 1.5 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.4 | 6.1 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.3 | 1.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.3 | 2.1 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.2 | 1.9 | REACTOME GAP JUNCTION TRAFFICKING | Genes involved in Gap junction trafficking |
| 0.2 | 2.6 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.2 | 1.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.2 | 3.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.2 | 3.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 1.8 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 1.0 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 1.5 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 1.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 1.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 2.0 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 0.9 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 1.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.9 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 2.0 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 7.1 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.7 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 1.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.4 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.2 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.9 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.8 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.4 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 0.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.5 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.3 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.1 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |