Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
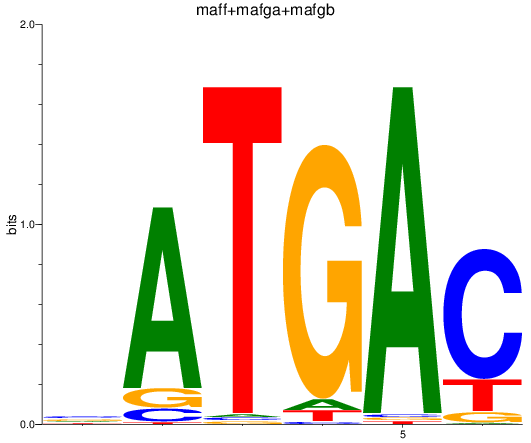
Results for maff+mafga+mafgb
Z-value: 1.02
Transcription factors associated with maff+mafga+mafgb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mafga
|
ENSDARG00000018109 | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Ga |
|
maff
|
ENSDARG00000028957 | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
|
mafgb
|
ENSDARG00000100097 | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Gb |
|
maff
|
ENSDARG00000111050 | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mafga | dr11_v1_chr3_-_55404985_55404985 | 0.56 | 1.2e-01 | Click! |
| mafgb | dr11_v1_chr11_+_45299447_45299447 | 0.15 | 7.1e-01 | Click! |
| maff | dr11_v1_chr12_-_19346678_19346678 | -0.02 | 9.6e-01 | Click! |
Activity profile of maff+mafga+mafgb motif
Sorted Z-values of maff+mafga+mafgb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_32526799 | 3.33 |
ENSDART00000185755
|
si:ch73-367p23.2
|
si:ch73-367p23.2 |
| chr3_-_55139127 | 1.98 |
ENSDART00000115324
|
hbae1.3
|
hemoglobin, alpha embryonic 1.3 |
| chr3_-_55128258 | 1.17 |
ENSDART00000101734
|
hbae1
|
hemoglobin, alpha embryonic 1 |
| chr11_+_30729745 | 1.17 |
ENSDART00000103270
|
slc22a7a
|
solute carrier family 22 (organic anion transporter), member 7a |
| chr3_+_33300522 | 0.91 |
ENSDART00000114023
|
hspb9
|
heat shock protein, alpha-crystallin-related, 9 |
| chr13_+_24280380 | 0.89 |
ENSDART00000184115
|
acta1b
|
actin, alpha 1b, skeletal muscle |
| chr3_-_41795917 | 0.84 |
ENSDART00000182662
|
grifin
|
galectin-related inter-fiber protein |
| chr18_+_808911 | 0.83 |
ENSDART00000172518
|
cox5ab
|
cytochrome c oxidase subunit Vab |
| chr23_-_28347039 | 0.83 |
ENSDART00000145072
|
neurod4
|
neuronal differentiation 4 |
| chr2_-_36040820 | 0.79 |
ENSDART00000003550
|
nmnat2
|
nicotinamide nucleotide adenylyltransferase 2 |
| chr19_-_19339285 | 0.74 |
ENSDART00000158413
ENSDART00000170479 |
cspg5b
|
chondroitin sulfate proteoglycan 5b |
| chr21_-_45613564 | 0.71 |
ENSDART00000160324
|
LO018363.1
|
|
| chr12_+_20336070 | 0.70 |
ENSDART00000066385
|
zgc:163057
|
zgc:163057 |
| chr21_-_26495700 | 0.69 |
ENSDART00000109379
|
cd248b
|
CD248 molecule, endosialin b |
| chr21_-_10773344 | 0.67 |
ENSDART00000063244
|
grp
|
gastrin-releasing peptide |
| chr24_-_32408404 | 0.66 |
ENSDART00000144157
|
si:ch211-56a11.2
|
si:ch211-56a11.2 |
| chr1_-_10048514 | 0.66 |
ENSDART00000125358
ENSDART00000054835 |
rnf175
|
ring finger protein 175 |
| chr7_-_24491614 | 0.65 |
ENSDART00000131063
|
si:dkeyp-75h12.5
|
si:dkeyp-75h12.5 |
| chr7_+_8543317 | 0.62 |
ENSDART00000114998
|
jac8
|
jacalin 8 |
| chr20_+_20637866 | 0.56 |
ENSDART00000060203
ENSDART00000079079 |
rtn1b
|
reticulon 1b |
| chr12_+_6041575 | 0.54 |
ENSDART00000091868
|
g6pca.2
|
glucose-6-phosphatase a, catalytic subunit, tandem duplicate 2 |
| chr19_+_25649626 | 0.54 |
ENSDART00000146947
|
tac1
|
tachykinin 1 |
| chr3_-_26109322 | 0.54 |
ENSDART00000113780
|
zgc:162612
|
zgc:162612 |
| chr8_-_54304381 | 0.53 |
ENSDART00000184177
|
RHO (1 of many)
|
rhodopsin |
| chr25_+_13191615 | 0.53 |
ENSDART00000168849
|
si:ch211-147m6.2
|
si:ch211-147m6.2 |
| chr19_-_1961024 | 0.53 |
ENSDART00000108784
|
mturn
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr4_+_3358383 | 0.53 |
ENSDART00000075320
|
nampta
|
nicotinamide phosphoribosyltransferase a |
| chr21_-_43606502 | 0.53 |
ENSDART00000151030
|
si:ch73-362m14.4
|
si:ch73-362m14.4 |
| chr9_-_23922011 | 0.53 |
ENSDART00000145734
|
col6a3
|
collagen, type VI, alpha 3 |
| chr7_+_25059845 | 0.51 |
ENSDART00000077215
|
ppp2r5b
|
protein phosphatase 2, regulatory subunit B', beta |
| chr16_+_10777116 | 0.50 |
ENSDART00000190902
|
atp1a3b
|
ATPase Na+/K+ transporting subunit alpha 3b |
| chr3_+_31933893 | 0.50 |
ENSDART00000146509
ENSDART00000139644 |
lin7b
|
lin-7 homolog B (C. elegans) |
| chr16_-_12173554 | 0.49 |
ENSDART00000110567
ENSDART00000155935 |
clstn3
|
calsyntenin 3 |
| chr24_+_9744012 | 0.47 |
ENSDART00000129656
|
tmem108
|
transmembrane protein 108 |
| chr8_-_31075015 | 0.46 |
ENSDART00000010993
|
slc20a1a
|
solute carrier family 20, member 1a |
| chr6_+_27667359 | 0.45 |
ENSDART00000159624
ENSDART00000049177 |
rab6ba
|
RAB6B, member RAS oncogene family a |
| chr21_+_27416284 | 0.44 |
ENSDART00000077593
ENSDART00000108763 |
cfb
|
complement factor B |
| chr10_-_26512742 | 0.44 |
ENSDART00000135951
|
si:dkey-5g14.1
|
si:dkey-5g14.1 |
| chr23_-_27345425 | 0.44 |
ENSDART00000022042
ENSDART00000191870 |
scn8aa
|
sodium channel, voltage gated, type VIII, alpha subunit a |
| chr17_+_12698532 | 0.44 |
ENSDART00000064509
ENSDART00000136830 |
stmn4l
|
stathmin-like 4, like |
| chr8_+_25351863 | 0.43 |
ENSDART00000034092
|
dnase1l1l
|
deoxyribonuclease I-like 1-like |
| chr9_-_4506819 | 0.43 |
ENSDART00000113975
|
kcnj3a
|
potassium inwardly-rectifying channel, subfamily J, member 3a |
| chr14_-_30490763 | 0.43 |
ENSDART00000193166
ENSDART00000183471 ENSDART00000087859 |
micu3b
|
mitochondrial calcium uptake family, member 3b |
| chr15_-_44512461 | 0.42 |
ENSDART00000155456
|
gria4a
|
glutamate receptor, ionotropic, AMPA 4a |
| chr10_+_21758811 | 0.42 |
ENSDART00000188827
|
pcdh1g11
|
protocadherin 1 gamma 11 |
| chr5_-_9073433 | 0.42 |
ENSDART00000099891
|
atp5meb
|
ATP synthase membrane subunit eb |
| chr20_+_20638034 | 0.41 |
ENSDART00000189759
|
rtn1b
|
reticulon 1b |
| chr12_+_48216662 | 0.41 |
ENSDART00000187369
|
lrrc20
|
leucine rich repeat containing 20 |
| chr12_-_10300101 | 0.41 |
ENSDART00000126428
|
mpp2b
|
membrane protein, palmitoylated 2b (MAGUK p55 subfamily member 2) |
| chr19_+_17385561 | 0.41 |
ENSDART00000141397
ENSDART00000143913 ENSDART00000133626 |
rpl15
|
ribosomal protein L15 |
| chr20_-_54259780 | 0.40 |
ENSDART00000172631
|
fkbp3
|
FK506 binding protein 3 |
| chr7_+_50766094 | 0.40 |
ENSDART00000165037
|
si:ch73-380l10.2
|
si:ch73-380l10.2 |
| chr10_-_26512993 | 0.39 |
ENSDART00000188549
ENSDART00000193316 |
si:dkey-5g14.1
|
si:dkey-5g14.1 |
| chr3_-_39171968 | 0.39 |
ENSDART00000154494
|
si:dkeyp-57f11.2
|
si:dkeyp-57f11.2 |
| chr17_-_31659670 | 0.38 |
ENSDART00000030448
|
vsx2
|
visual system homeobox 2 |
| chr2_-_24996441 | 0.38 |
ENSDART00000144795
|
slc35g2a
|
solute carrier family 35, member G2a |
| chr21_-_16114061 | 0.38 |
ENSDART00000035742
|
cyb561a3b
|
cytochrome b561 family, member A3b |
| chr23_-_26535875 | 0.38 |
ENSDART00000135988
|
si:dkey-205h13.2
|
si:dkey-205h13.2 |
| chr5_-_7897316 | 0.38 |
ENSDART00000160743
ENSDART00000160912 |
opn4xb
|
opsin 4xb |
| chr10_-_33621739 | 0.37 |
ENSDART00000142655
ENSDART00000128049 |
hunk
|
hormonally up-regulated Neu-associated kinase |
| chr3_+_15907458 | 0.37 |
ENSDART00000163525
|
mapk8ip3
|
mitogen-activated protein kinase 8 interacting protein 3 |
| chr23_-_4915118 | 0.36 |
ENSDART00000060714
|
atp6ap1a
|
ATPase H+ transporting accessory protein 1a |
| chr23_+_216012 | 0.36 |
ENSDART00000181115
ENSDART00000004678 ENSDART00000190439 ENSDART00000189322 |
PDZD4
|
si:ch73-162j3.4 |
| chr5_-_63509581 | 0.36 |
ENSDART00000097325
|
c5
|
complement component 5 |
| chr13_+_36146415 | 0.36 |
ENSDART00000140301
|
TTC9
|
si:ch211-259k16.3 |
| chr8_+_39607466 | 0.36 |
ENSDART00000097427
|
msi1
|
musashi RNA-binding protein 1 |
| chr6_+_584632 | 0.36 |
ENSDART00000151150
|
zgc:92360
|
zgc:92360 |
| chr7_+_25036188 | 0.35 |
ENSDART00000163957
ENSDART00000169749 |
sb:cb1058
|
sb:cb1058 |
| chr19_-_17385548 | 0.35 |
ENSDART00000162383
|
nkiras1
|
NFKB inhibitor interacting Ras-like 1 |
| chr21_-_41147818 | 0.35 |
ENSDART00000167339
ENSDART00000192730 |
msx2b
|
muscle segment homeobox 2b |
| chr19_+_43563179 | 0.35 |
ENSDART00000151478
|
cd164l2
|
CD164 sialomucin-like 2 |
| chr14_-_17072736 | 0.34 |
ENSDART00000106333
|
phox2bb
|
paired-like homeobox 2bb |
| chr7_+_42206543 | 0.34 |
ENSDART00000112543
|
phkb
|
phosphorylase kinase, beta |
| chr10_+_20113830 | 0.33 |
ENSDART00000139722
|
dmtn
|
dematin actin binding protein |
| chr16_+_12240605 | 0.33 |
ENSDART00000060056
|
tpi1b
|
triosephosphate isomerase 1b |
| chr25_+_459901 | 0.33 |
ENSDART00000154895
|
prtgb
|
protogenin homolog b (Gallus gallus) |
| chr6_-_46589726 | 0.33 |
ENSDART00000084334
|
ptgis
|
prostaglandin I2 (prostacyclin) synthase |
| chr15_-_34418525 | 0.32 |
ENSDART00000147582
|
agmo
|
alkylglycerol monooxygenase |
| chr7_-_57933736 | 0.32 |
ENSDART00000142580
|
ank2b
|
ankyrin 2b, neuronal |
| chr6_+_52064304 | 0.32 |
ENSDART00000153468
|
abraa
|
actin binding Rho activating protein a |
| chr24_+_38301080 | 0.32 |
ENSDART00000105672
|
mybpc2b
|
myosin binding protein C, fast type b |
| chr6_+_120181 | 0.31 |
ENSDART00000151209
ENSDART00000185930 |
cdkn2d
|
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) |
| chr24_-_25428176 | 0.31 |
ENSDART00000090010
|
phex
|
phosphate regulating endopeptidase homolog, X-linked |
| chr9_+_27411502 | 0.31 |
ENSDART00000143994
|
si:dkey-193n17.9
|
si:dkey-193n17.9 |
| chr7_-_35516251 | 0.31 |
ENSDART00000045628
|
irx6a
|
iroquois homeobox 6a |
| chr20_+_2281933 | 0.31 |
ENSDART00000137579
|
si:ch73-18b11.2
|
si:ch73-18b11.2 |
| chr22_+_1796057 | 0.31 |
ENSDART00000170834
|
znf1179
|
zinc finger protein 1179 |
| chr25_+_20077225 | 0.30 |
ENSDART00000136543
|
tnni4b.1
|
troponin I4b, tandem duplicate 1 |
| chr17_-_24564674 | 0.30 |
ENSDART00000105435
ENSDART00000135086 |
abch1
|
ATP-binding cassette, sub-family H, member 1 |
| chr10_-_27049170 | 0.29 |
ENSDART00000143451
|
cnih2
|
cornichon family AMPA receptor auxiliary protein 2 |
| chr21_+_9666347 | 0.29 |
ENSDART00000163853
|
mapk10
|
mitogen-activated protein kinase 10 |
| chr11_+_25112269 | 0.29 |
ENSDART00000147546
|
ndrg3a
|
ndrg family member 3a |
| chr6_-_35446110 | 0.28 |
ENSDART00000058773
|
rgs16
|
regulator of G protein signaling 16 |
| chr3_-_32603191 | 0.28 |
ENSDART00000150997
|
si:ch73-248e21.7
|
si:ch73-248e21.7 |
| chr18_+_48446704 | 0.28 |
ENSDART00000134817
|
fli1a
|
Fli-1 proto-oncogene, ETS transcription factor a |
| chr21_-_28340977 | 0.28 |
ENSDART00000141629
|
nrxn2a
|
neurexin 2a |
| chr4_-_17805128 | 0.28 |
ENSDART00000128988
|
spi2
|
Spi-2 proto-oncogene |
| chr22_-_11136625 | 0.28 |
ENSDART00000016873
ENSDART00000125561 |
atp6ap2
|
ATPase H+ transporting accessory protein 2 |
| chr9_-_18424844 | 0.28 |
ENSDART00000154351
|
enox1
|
ecto-NOX disulfide-thiol exchanger 1 |
| chr19_+_46372115 | 0.27 |
ENSDART00000163935
|
med30
|
mediator complex subunit 30 |
| chr19_+_32947910 | 0.27 |
ENSDART00000052091
|
atp6v1c1b
|
ATPase H+ transporting V1 subunit C1b |
| chr21_-_22928214 | 0.27 |
ENSDART00000182760
|
dub
|
duboraya |
| chr1_-_45157243 | 0.27 |
ENSDART00000131882
|
mucms1
|
mucin, multiple PTS and SEA group, member 1 |
| chr3_+_32403758 | 0.26 |
ENSDART00000156982
|
si:ch211-195b15.8
|
si:ch211-195b15.8 |
| chr7_+_11543999 | 0.26 |
ENSDART00000173676
|
il16
|
interleukin 16 |
| chr23_-_26536055 | 0.26 |
ENSDART00000182719
|
si:dkey-205h13.2
|
si:dkey-205h13.2 |
| chr10_-_27009413 | 0.26 |
ENSDART00000139942
ENSDART00000146983 ENSDART00000132352 |
uqcc3
|
ubiquinol-cytochrome c reductase complex assembly factor 3 |
| chr7_+_69653981 | 0.25 |
ENSDART00000090165
|
frem1a
|
Fras1 related extracellular matrix 1a |
| chr5_+_23118470 | 0.25 |
ENSDART00000149893
|
nexmifa
|
neurite extension and migration factor a |
| chr18_+_2222447 | 0.25 |
ENSDART00000185927
|
pigbos1
|
PIGB opposite strand 1 |
| chr5_+_53824959 | 0.24 |
ENSDART00000169565
|
sat2b
|
spermidine/spermine N1-acetyltransferase family member 2b |
| chr6_-_48087152 | 0.24 |
ENSDART00000180614
|
slc2a1b
|
solute carrier family 2 (facilitated glucose transporter), member 1b |
| chr11_+_37216668 | 0.24 |
ENSDART00000173076
|
zgc:112265
|
zgc:112265 |
| chr16_-_29452039 | 0.24 |
ENSDART00000148960
|
si:ch211-113g11.6
|
si:ch211-113g11.6 |
| chr16_+_26777473 | 0.24 |
ENSDART00000188870
|
cdh17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr23_+_19675780 | 0.24 |
ENSDART00000124335
|
smim4
|
small integral membrane protein 4 |
| chr3_-_22829710 | 0.24 |
ENSDART00000055659
|
cyb561
|
cytochrome b561 |
| chr17_-_22062364 | 0.23 |
ENSDART00000114470
|
ttbk1b
|
tau tubulin kinase 1b |
| chr1_+_11107688 | 0.23 |
ENSDART00000109858
|
knstrn
|
kinetochore-localized astrin/SPAG5 binding protein |
| chr6_-_10780698 | 0.23 |
ENSDART00000151714
|
gpr155b
|
G protein-coupled receptor 155b |
| chr3_-_57744323 | 0.23 |
ENSDART00000101829
|
lgals3bpb
|
lectin, galactoside-binding, soluble, 3 binding protein b |
| chr17_+_27134806 | 0.23 |
ENSDART00000151901
|
rps6ka1
|
ribosomal protein S6 kinase a, polypeptide 1 |
| chr19_-_13286722 | 0.23 |
ENSDART00000168296
ENSDART00000158330 |
zfpm2b
|
zinc finger protein, FOG family member 2b |
| chr3_+_41647637 | 0.23 |
ENSDART00000050332
|
gna12a
|
guanine nucleotide binding protein (G protein) alpha 12a |
| chr20_+_32224405 | 0.22 |
ENSDART00000062993
ENSDART00000147448 |
sesn1
|
sestrin 1 |
| chr9_-_42861080 | 0.22 |
ENSDART00000193688
|
ttn.1
|
titin, tandem duplicate 1 |
| chr3_-_32320537 | 0.22 |
ENSDART00000113550
ENSDART00000168483 |
si:dkey-16p21.7
|
si:dkey-16p21.7 |
| chr14_+_25464681 | 0.22 |
ENSDART00000067500
ENSDART00000187601 |
si:dkey-280e21.3
|
si:dkey-280e21.3 |
| chr11_-_29658396 | 0.22 |
ENSDART00000183947
|
rpl22
|
ribosomal protein L22 |
| chr5_-_63286077 | 0.21 |
ENSDART00000131274
|
tbcelb
|
tubulin folding cofactor E-like b |
| chr7_+_34290051 | 0.21 |
ENSDART00000123498
|
fibinb
|
fin bud initiation factor b |
| chr7_+_42208859 | 0.21 |
ENSDART00000148643
|
phkb
|
phosphorylase kinase, beta |
| chr18_-_49283058 | 0.21 |
ENSDART00000076554
|
si:zfos-464b6.2
|
si:zfos-464b6.2 |
| chr7_-_18730474 | 0.21 |
ENSDART00000170374
|
si:dkey-38l22.2
|
si:dkey-38l22.2 |
| chr11_+_329687 | 0.21 |
ENSDART00000172882
|
cyp27b1
|
cytochrome P450, family 27, subfamily B, polypeptide 1 |
| chr7_-_20338048 | 0.21 |
ENSDART00000125594
|
zgc:194312
|
zgc:194312 |
| chr5_-_41307550 | 0.20 |
ENSDART00000143446
|
npr3
|
natriuretic peptide receptor 3 |
| chr1_+_26411496 | 0.20 |
ENSDART00000112263
|
arhgef38
|
Rho guanine nucleotide exchange factor (GEF) 38 |
| chr9_-_21230999 | 0.20 |
ENSDART00000150160
ENSDART00000102147 |
pla1a
|
phospholipase A1 member A |
| chr24_-_982443 | 0.20 |
ENSDART00000063151
|
napga
|
N-ethylmaleimide-sensitive factor attachment protein, gamma a |
| chr14_+_39255437 | 0.20 |
ENSDART00000147443
|
diaph2
|
diaphanous-related formin 2 |
| chr21_+_9576176 | 0.20 |
ENSDART00000161289
ENSDART00000159899 ENSDART00000162834 |
mapk10
|
mitogen-activated protein kinase 10 |
| chr6_+_34512313 | 0.20 |
ENSDART00000102554
|
lrp8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr18_-_15559817 | 0.19 |
ENSDART00000061681
|
si:ch211-245j22.3
|
si:ch211-245j22.3 |
| chr19_-_25519310 | 0.19 |
ENSDART00000089882
|
C1GALT1 (1 of many)
|
si:dkey-202e17.1 |
| chr1_-_59104145 | 0.19 |
ENSDART00000132495
ENSDART00000152457 |
MFAP4 (1 of many)
si:zfos-2330d3.7
|
si:zfos-2330d3.1 si:zfos-2330d3.7 |
| chr22_-_32392252 | 0.19 |
ENSDART00000148681
ENSDART00000137945 |
pcbp4
|
poly(rC) binding protein 4 |
| chr8_+_14058646 | 0.19 |
ENSDART00000080852
|
ugt5e1
|
UDP glucuronosyltransferase 5 family, polypeptide E1 |
| chr13_-_42560662 | 0.19 |
ENSDART00000124898
|
CR792417.1
|
|
| chr2_+_23222939 | 0.18 |
ENSDART00000026800
|
kifap3b
|
kinesin-associated protein 3b |
| chr20_+_46183505 | 0.18 |
ENSDART00000060799
|
taar13b
|
trace amine associated receptor 13b |
| chr6_-_31348999 | 0.18 |
ENSDART00000153734
|
dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr5_+_20589472 | 0.18 |
ENSDART00000130911
|
cmklr1
|
chemokine-like receptor 1 |
| chr12_-_48960308 | 0.18 |
ENSDART00000176247
|
CABZ01112647.1
|
|
| chr9_+_28688574 | 0.18 |
ENSDART00000101319
|
zgc:162396
|
zgc:162396 |
| chr25_+_18711804 | 0.18 |
ENSDART00000011149
|
fam185a
|
family with sequence similarity 185, member A |
| chr7_+_26709251 | 0.18 |
ENSDART00000149426
ENSDART00000010323 |
cd82a
|
CD82 molecule a |
| chr20_+_52546186 | 0.18 |
ENSDART00000110777
ENSDART00000153377 ENSDART00000153013 ENSDART00000042704 |
eef1db
|
eukaryotic translation elongation factor 1 delta b (guanine nucleotide exchange protein) |
| chr10_+_29698467 | 0.17 |
ENSDART00000163402
|
dlg2
|
discs, large homolog 2 (Drosophila) |
| chr18_-_8579907 | 0.17 |
ENSDART00000147284
|
FRMD4A
|
si:ch211-220f12.1 |
| chr23_+_44634187 | 0.17 |
ENSDART00000143688
|
si:ch73-265d7.2
|
si:ch73-265d7.2 |
| chr2_-_32505091 | 0.17 |
ENSDART00000141884
ENSDART00000056639 |
faim2a
|
Fas apoptotic inhibitory molecule 2a |
| chr10_-_167782 | 0.17 |
ENSDART00000108780
|
erg
|
v-ets avian erythroblastosis virus E26 oncogene homolog |
| chr11_-_29936755 | 0.17 |
ENSDART00000145251
|
pir
|
pirin |
| chr3_+_24189804 | 0.17 |
ENSDART00000134723
|
prr15la
|
proline rich 15-like a |
| chr24_-_17029374 | 0.17 |
ENSDART00000039267
|
ptgdsb.1
|
prostaglandin D2 synthase b, tandem duplicate 1 |
| chr11_-_41130239 | 0.16 |
ENSDART00000173268
|
dnajc11b
|
DnaJ (Hsp40) homolog, subfamily C, member 11b |
| chr6_-_49898881 | 0.16 |
ENSDART00000150204
|
atp5f1e
|
ATP synthase F1 subunit epsilon |
| chr10_-_25699454 | 0.16 |
ENSDART00000064376
|
sod1
|
superoxide dismutase 1, soluble |
| chr14_-_2355833 | 0.16 |
ENSDART00000157677
|
PCDHGC3 (1 of many)
|
si:ch73-233f7.6 |
| chr2_-_9818640 | 0.16 |
ENSDART00000139499
ENSDART00000165548 ENSDART00000012442 ENSDART00000046587 |
ap2m1b
|
adaptor-related protein complex 2, mu 1 subunit, b |
| chr18_-_49116382 | 0.16 |
ENSDART00000174157
|
BX663503.3
|
|
| chr8_+_25352268 | 0.16 |
ENSDART00000187829
|
dnase1l1l
|
deoxyribonuclease I-like 1-like |
| chr20_+_26538137 | 0.15 |
ENSDART00000045397
|
stx11b.1
|
syntaxin 11b, tandem duplicate 1 |
| chr5_-_26950374 | 0.15 |
ENSDART00000050542
|
htra4
|
HtrA serine peptidase 4 |
| chr2_+_36862473 | 0.15 |
ENSDART00000135624
|
si:dkey-193b15.8
|
si:dkey-193b15.8 |
| chr14_+_33413980 | 0.15 |
ENSDART00000052780
ENSDART00000124437 ENSDART00000173327 |
ndufa1
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 1 |
| chr16_-_22192006 | 0.15 |
ENSDART00000163338
|
il6r
|
interleukin 6 receptor |
| chr23_-_29502287 | 0.15 |
ENSDART00000141075
ENSDART00000053807 |
kif1b
|
kinesin family member 1B |
| chr6_-_51101834 | 0.15 |
ENSDART00000092493
|
ptprt
|
protein tyrosine phosphatase, receptor type, t |
| chr18_+_8231138 | 0.15 |
ENSDART00000140193
|
arsa
|
arylsulfatase A |
| chr19_-_8962884 | 0.15 |
ENSDART00000172582
ENSDART00000104657 |
mrps21
|
mitochondrial ribosomal protein S21 |
| chr2_-_9816492 | 0.15 |
ENSDART00000138472
ENSDART00000124051 |
ap2m1b
|
adaptor-related protein complex 2, mu 1 subunit, b |
| chr8_-_25327809 | 0.15 |
ENSDART00000137242
|
eps8l3b
|
EPS8-like 3b |
| chr2_-_32356539 | 0.14 |
ENSDART00000169316
|
asap1a
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1a |
| chr7_-_5316901 | 0.14 |
ENSDART00000181505
ENSDART00000124367 |
si:cabz01074946.1
|
si:cabz01074946.1 |
| chr7_+_54617060 | 0.14 |
ENSDART00000158898
|
fgf4
|
fibroblast growth factor 4 |
| chr10_-_41664427 | 0.14 |
ENSDART00000150213
|
ggt1b
|
gamma-glutamyltransferase 1b |
| chr8_+_53452681 | 0.14 |
ENSDART00000166705
|
cacna1db
|
calcium channel, voltage-dependent, L type, alpha 1D subunit, b |
| chr4_+_25693463 | 0.14 |
ENSDART00000132864
|
acot18
|
acyl-CoA thioesterase 18 |
| chr20_-_27864964 | 0.14 |
ENSDART00000153311
|
syndig1l
|
synapse differentiation inducing 1-like |
| chr24_+_926258 | 0.14 |
ENSDART00000153654
|
piezo2a.2
|
piezo-type mechanosensitive ion channel component 2a, tandem duplicate 2 |
| chr16_+_42829735 | 0.13 |
ENSDART00000014956
|
polr3glb
|
polymerase (RNA) III (DNA directed) polypeptide G like b |
| chr11_-_22372072 | 0.13 |
ENSDART00000065996
|
tmem183a
|
transmembrane protein 183A |
| chr17_+_24006792 | 0.13 |
ENSDART00000122415
|
si:ch211-63b16.4
|
si:ch211-63b16.4 |
| chr6_+_9163007 | 0.13 |
ENSDART00000183054
ENSDART00000157552 |
zgc:112023
|
zgc:112023 |
| chr25_+_32474031 | 0.13 |
ENSDART00000152124
|
sqor
|
sulfide quinone oxidoreductase |
| chr2_+_30721466 | 0.13 |
ENSDART00000128982
|
si:dkey-94e7.2
|
si:dkey-94e7.2 |
| chr17_+_45009868 | 0.13 |
ENSDART00000085009
|
coq6
|
coenzyme Q6 monooxygenase |
| chr8_-_40075983 | 0.13 |
ENSDART00000141455
|
ggt1a
|
gamma-glutamyltransferase 1a |
Network of associatons between targets according to the STRING database.
First level regulatory network of maff+mafga+mafgb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.7 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 0.8 | GO:0001839 | neural plate morphogenesis(GO:0001839) |
| 0.1 | 0.5 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.1 | 0.4 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.1 | 0.5 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.1 | 0.5 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.1 | 0.5 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 | 0.3 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) |
| 0.1 | 0.9 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.1 | 0.3 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.1 | 0.3 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 | 0.7 | GO:0099550 | trans-synaptic signalling, modulating synaptic transmission(GO:0099550) |
| 0.1 | 0.3 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 0.8 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.3 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.8 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.1 | 0.3 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.1 | 0.5 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 0.3 | GO:0031179 | peptide modification(GO:0031179) |
| 0.0 | 0.1 | GO:0048917 | posterior lateral line ganglion development(GO:0048917) |
| 0.0 | 0.2 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.1 | GO:0070221 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.0 | 0.3 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.2 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.0 | 0.1 | GO:0045040 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.2 | GO:0048025 | negative regulation of RNA splicing(GO:0033119) negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.1 | GO:1901232 | regulation of convergent extension involved in axis elongation(GO:1901232) positive regulation of epiboly involved in gastrulation with mouth forming second(GO:1904088) |
| 0.0 | 0.2 | GO:0098529 | neuromuscular junction development, skeletal muscle fiber(GO:0098529) |
| 0.0 | 0.2 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.6 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.0 | 0.2 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 | 0.1 | GO:0010312 | detoxification of zinc ion(GO:0010312) stress response to zinc ion(GO:1990359) |
| 0.0 | 0.2 | GO:1901031 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.0 | 0.4 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.5 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0009838 | abscission(GO:0009838) mitotic cytokinesis checkpoint(GO:0044878) |
| 0.0 | 0.1 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.0 | 0.4 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.5 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 0.1 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.6 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.2 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.2 | GO:0048662 | regulation of smooth muscle cell proliferation(GO:0048660) negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.0 | 0.1 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.3 | GO:0046457 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 | 0.1 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.0 | 0.3 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.4 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.0 | 0.5 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.2 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.0 | 0.3 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.0 | 0.1 | GO:0071467 | cellular response to pH(GO:0071467) |
| 0.0 | 0.6 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.1 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.7 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 0.2 | GO:1990748 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.6 | GO:0010921 | regulation of phosphatase activity(GO:0010921) |
| 0.0 | 0.2 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.0 | 0.2 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.7 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.1 | GO:0003321 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) positive regulation of heart rate(GO:0010460) |
| 0.0 | 0.3 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.3 | GO:0050927 | regulation of positive chemotaxis(GO:0050926) positive regulation of positive chemotaxis(GO:0050927) induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.2 | GO:0071594 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.0 | 0.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.2 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.9 | GO:0048741 | myotube cell development(GO:0014904) skeletal muscle fiber development(GO:0048741) |
| 0.0 | 0.1 | GO:1903523 | negative regulation of heart contraction(GO:0045822) negative regulation of blood circulation(GO:1903523) |
| 0.0 | 0.1 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.0 | 0.8 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.3 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.0 | GO:0070285 | pigment cell development(GO:0070285) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.7 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 0.5 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 0.3 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.1 | 0.4 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.1 | 0.8 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.9 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 0.6 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.2 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.3 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.4 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.1 | GO:0008352 | katanin complex(GO:0008352) |
| 0.0 | 0.3 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.2 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.4 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.3 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.2 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.2 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.3 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.4 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.1 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.0 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.1 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.0 | 0.4 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.1 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.4 | GO:0005861 | troponin complex(GO:0005861) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.2 | 0.7 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.2 | 2.7 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.2 | 0.5 | GO:0031834 | neurokinin receptor binding(GO:0031834) substance P receptor binding(GO:0031835) |
| 0.1 | 0.5 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 0.3 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 0.4 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 0.3 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.1 | 0.2 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.1 | 0.2 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.1 | 0.7 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.6 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.1 | 0.3 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 0.5 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.4 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.5 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.4 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.3 | GO:0036374 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.8 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.1 | GO:0070224 | sulfide:quinone oxidoreductase activity(GO:0070224) |
| 0.0 | 0.1 | GO:0005183 | gonadotropin hormone-releasing hormone activity(GO:0005183) gonadotropin-releasing hormone receptor binding(GO:0031530) |
| 0.0 | 0.3 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.2 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.0 | 0.2 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.2 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.5 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.8 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.0 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.0 | 0.4 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.5 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.1 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.1 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.2 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.3 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.2 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.2 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.4 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.2 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.3 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.0 | GO:0008397 | sterol 12-alpha-hydroxylase activity(GO:0008397) |
| 0.0 | 0.0 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.0 | 0.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.2 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.3 | GO:0046961 | ATPase activity, coupled to transmembrane movement of ions, rotational mechanism(GO:0044769) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.4 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.2 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 0.7 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.1 | 0.7 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.5 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.6 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.1 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.5 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.2 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.0 | 0.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.4 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.1 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |