Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
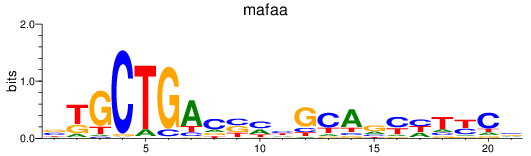
Results for mafaa
Z-value: 1.22
Transcription factors associated with mafaa
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mafaa
|
ENSDARG00000044155 | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Aa |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mafaa | dr11_v1_chr6_-_32703317_32703317 | 0.74 | 2.2e-02 | Click! |
Activity profile of mafaa motif
Sorted Z-values of mafaa motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_39313027 | 5.97 |
ENSDART00000012644
|
krt4
|
keratin 4 |
| chr13_+_4671698 | 5.04 |
ENSDART00000164617
ENSDART00000128494 ENSDART00000165776 |
pla2g12b
|
phospholipase A2, group XIIB |
| chr24_-_41312459 | 3.25 |
ENSDART00000041349
|
crygn2
|
crystallin, gamma N2 |
| chr8_+_35172594 | 2.78 |
ENSDART00000177146
|
BX897670.1
|
|
| chr16_+_1353894 | 2.41 |
ENSDART00000148426
|
celf3b
|
cugbp, Elav-like family member 3b |
| chr19_-_5332784 | 2.38 |
ENSDART00000010373
|
krt1-19d
|
keratin, type 1, gene 19d |
| chr10_+_21780250 | 2.15 |
ENSDART00000183782
|
pcdh1g9
|
protocadherin 1 gamma 9 |
| chr23_-_21453614 | 2.13 |
ENSDART00000079274
|
her4.1
|
hairy-related 4, tandem duplicate 1 |
| chr5_-_40510397 | 2.05 |
ENSDART00000146237
ENSDART00000051065 |
fsta
|
follistatin a |
| chr18_-_48745517 | 1.98 |
ENSDART00000097259
|
st3gal4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr23_+_21459263 | 1.93 |
ENSDART00000104209
|
her4.3
|
hairy-related 4, tandem duplicate 3 |
| chr1_-_625875 | 1.82 |
ENSDART00000167331
|
appa
|
amyloid beta (A4) precursor protein a |
| chr12_-_47558276 | 1.62 |
ENSDART00000160260
|
grem2b
|
gremlin 2, DAN family BMP antagonist b |
| chr1_+_52392511 | 1.60 |
ENSDART00000144025
|
si:ch211-217k17.8
|
si:ch211-217k17.8 |
| chr22_-_27709692 | 1.42 |
ENSDART00000172458
|
CR547131.1
|
|
| chr7_+_6969909 | 1.42 |
ENSDART00000189886
|
actn3b
|
actinin alpha 3b |
| chr23_+_42338325 | 1.34 |
ENSDART00000169660
|
cyp2aa7
|
cytochrome P450, family 2, subfamily AA, polypeptide 7 |
| chr19_-_28788466 | 1.29 |
ENSDART00000151793
|
sox4a
|
SRY (sex determining region Y)-box 4a |
| chr21_+_45841731 | 1.28 |
ENSDART00000038657
|
faxdc2
|
fatty acid hydroxylase domain containing 2 |
| chr2_+_38161318 | 1.28 |
ENSDART00000044264
|
mmp14b
|
matrix metallopeptidase 14b (membrane-inserted) |
| chr23_+_2136344 | 1.23 |
ENSDART00000085290
ENSDART00000092757 |
cpeb2
|
cytoplasmic polyadenylation element binding protein 2 |
| chr15_+_7086327 | 1.22 |
ENSDART00000114560
|
pik3cb
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit beta |
| chr16_+_4658250 | 1.22 |
ENSDART00000006212
|
LIN28A
|
si:ch1073-284b18.2 |
| chr4_+_25950372 | 1.13 |
ENSDART00000125767
|
metap2a
|
methionyl aminopeptidase 2a |
| chr19_+_29798064 | 1.09 |
ENSDART00000167803
ENSDART00000051804 |
marcksl1b
|
MARCKS-like 1b |
| chr11_-_36263886 | 0.99 |
ENSDART00000140397
|
nfya
|
nuclear transcription factor Y, alpha |
| chr23_+_3538463 | 0.97 |
ENSDART00000172758
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr19_-_42566527 | 0.95 |
ENSDART00000147818
ENSDART00000142135 |
si:dkey-228a15.1
|
si:dkey-228a15.1 |
| chr19_-_48010490 | 0.95 |
ENSDART00000159938
|
FBXL19
|
zgc:158376 |
| chr2_-_37896965 | 0.85 |
ENSDART00000129852
|
hbl1
|
hexose-binding lectin 1 |
| chr1_+_59090743 | 0.82 |
ENSDART00000100199
|
mfap4
|
microfibril associated protein 4 |
| chr1_+_59090972 | 0.74 |
ENSDART00000171497
|
mfap4
|
microfibril associated protein 4 |
| chr14_-_17588345 | 0.74 |
ENSDART00000143486
|
selenot2
|
selenoprotein T, 2 |
| chr23_+_12916062 | 0.73 |
ENSDART00000144268
|
si:dkey-150i13.2
|
si:dkey-150i13.2 |
| chr24_+_80653 | 0.70 |
ENSDART00000158473
ENSDART00000129135 |
reck
|
reversion-inducing-cysteine-rich protein with kazal motifs |
| chr20_-_22476255 | 0.66 |
ENSDART00000103510
|
pdgfra
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr8_-_2591654 | 0.64 |
ENSDART00000049109
|
seta
|
SET nuclear proto-oncogene a |
| chr17_-_48915427 | 0.64 |
ENSDART00000054781
|
lgals8b
|
galectin 8b |
| chr2_-_22688651 | 0.62 |
ENSDART00000013863
|
agxtb
|
alanine-glyoxylate aminotransferase b |
| chr4_-_58448785 | 0.52 |
ENSDART00000172359
|
si:ch211-212k5.3
|
si:ch211-212k5.3 |
| chr6_-_131401 | 0.51 |
ENSDART00000151251
|
LRRC8E
|
si:zfos-323e3.4 |
| chr5_-_5326010 | 0.50 |
ENSDART00000161946
|
pbx3a
|
pre-B-cell leukemia homeobox 3a |
| chr22_+_21305682 | 0.46 |
ENSDART00000023521
|
odf3l2
|
outer dense fiber of sperm tails 3-like 2 |
| chr25_+_459901 | 0.45 |
ENSDART00000154895
|
prtgb
|
protogenin homolog b (Gallus gallus) |
| chr24_-_6029314 | 0.45 |
ENSDART00000136155
|
ftr60
|
finTRIM family, member 60 |
| chr5_+_51102010 | 0.41 |
ENSDART00000110377
|
zgc:194398
|
zgc:194398 |
| chr22_-_15280638 | 0.40 |
ENSDART00000063008
|
mfng
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr3_+_26322596 | 0.40 |
ENSDART00000172700
|
si:ch211-156b7.5
|
si:ch211-156b7.5 |
| chr18_+_33591704 | 0.40 |
ENSDART00000185631
ENSDART00000057845 |
si:dkey-47k20.5
|
si:dkey-47k20.5 |
| chr25_-_27842654 | 0.40 |
ENSDART00000154852
ENSDART00000156906 |
asb15a
|
ankyrin repeat and SOCS box containing 15a |
| chr14_+_40641350 | 0.38 |
ENSDART00000074506
|
tnmd
|
tenomodulin |
| chr15_-_18432673 | 0.35 |
ENSDART00000146853
|
ncam1b
|
neural cell adhesion molecule 1b |
| chr22_+_635813 | 0.32 |
ENSDART00000179067
|
CU856139.1
|
|
| chr18_+_45676788 | 0.31 |
ENSDART00000109948
ENSDART00000179864 |
qser1
|
glutamine and serine rich 1 |
| chr13_-_28272299 | 0.28 |
ENSDART00000006393
|
tlx1
|
T cell leukemia homeobox 1 |
| chr7_-_7493758 | 0.26 |
ENSDART00000036703
|
pfdn2
|
prefoldin subunit 2 |
| chr14_-_2004291 | 0.25 |
ENSDART00000114039
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr7_-_20756013 | 0.24 |
ENSDART00000185259
|
chd3
|
chromodomain helicase DNA binding protein 3 |
| chr3_-_1388936 | 0.24 |
ENSDART00000171278
|
ddx47
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 47 |
| chr14_-_48348973 | 0.23 |
ENSDART00000185822
|
CABZ01080056.1
|
|
| chr3_-_10677890 | 0.22 |
ENSDART00000155382
ENSDART00000171319 |
si:ch1073-144j5.2
|
si:ch1073-144j5.2 |
| chr4_-_8030583 | 0.22 |
ENSDART00000113628
|
si:ch211-240l19.8
|
si:ch211-240l19.8 |
| chr11_+_12719944 | 0.22 |
ENSDART00000054837
ENSDART00000144954 |
ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr3_-_18274691 | 0.22 |
ENSDART00000161140
|
BX649434.3
|
|
| chr4_+_19700308 | 0.21 |
ENSDART00000027919
|
pax4
|
paired box 4 |
| chr18_-_26126258 | 0.20 |
ENSDART00000145875
|
ANKRD34C
|
ankyrin repeat domain 34C |
| chr16_-_27676123 | 0.17 |
ENSDART00000180804
|
tbrg4
|
transforming growth factor beta regulator 4 |
| chr2_-_37465517 | 0.17 |
ENSDART00000139983
|
si:dkey-57k2.6
|
si:dkey-57k2.6 |
| chr22_+_28328156 | 0.15 |
ENSDART00000166714
|
impg2b
|
interphotoreceptor matrix proteoglycan 2b |
| chr5_-_25123807 | 0.15 |
ENSDART00000183171
|
abca2
|
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr1_-_53504603 | 0.14 |
ENSDART00000162025
ENSDART00000100788 |
commd1
|
copper metabolism (Murr1) domain containing 1 |
| chr8_-_14052349 | 0.13 |
ENSDART00000135811
|
atp2b3a
|
ATPase plasma membrane Ca2+ transporting 3a |
| chr25_+_31264155 | 0.12 |
ENSDART00000012256
|
tnni2a.3
|
troponin I type 2a (skeletal, fast), tandem duplicate 3 |
| chr4_-_72468168 | 0.10 |
ENSDART00000182995
ENSDART00000174067 |
CR788316.1
|
|
| chr1_+_59090583 | 0.10 |
ENSDART00000150658
|
mfap4
|
microfibril associated protein 4 |
| chr11_+_25276748 | 0.09 |
ENSDART00000126211
|
cyldb
|
cylindromatosis (turban tumor syndrome), b |
| chr20_-_23440955 | 0.08 |
ENSDART00000153386
|
slc10a4
|
solute carrier family 10, member 4 |
| chr12_-_11649690 | 0.08 |
ENSDART00000149713
|
btbd16
|
BTB (POZ) domain containing 16 |
| chr24_+_37709191 | 0.06 |
ENSDART00000066558
|
decr2
|
2,4-dienoyl CoA reductase 2, peroxisomal |
| chr9_+_1251514 | 0.06 |
ENSDART00000184806
ENSDART00000186356 ENSDART00000185683 |
FO818722.1
|
|
| chr21_+_19347655 | 0.05 |
ENSDART00000093155
|
hpse
|
heparanase |
| chr8_+_52479026 | 0.03 |
ENSDART00000162885
|
FO704661.1
|
Danio rerio gamma-glutamyl hydrolase (LOC553228), mRNA. |
| chr24_-_38079261 | 0.03 |
ENSDART00000105662
|
crp1
|
C-reactive protein 1 |
| chr20_-_39219537 | 0.02 |
ENSDART00000005764
|
cyp39a1
|
cytochrome P450, family 39, subfamily A, polypeptide 1 |
| chr25_+_3217419 | 0.01 |
ENSDART00000104859
|
rccd1
|
RCC1 domain containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of mafaa
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.0 | GO:0050482 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) arachidonate transport(GO:1903963) |
| 0.3 | 1.6 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.3 | 1.1 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 0.2 | 0.7 | GO:1905048 | regulation of metallopeptidase activity(GO:1905048) |
| 0.2 | 2.0 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.2 | 4.1 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.1 | 1.3 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 | 0.7 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.1 | 0.5 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 0.6 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.1 | 1.7 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 1.2 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 0.4 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 2.4 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 1.2 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.1 | GO:2000009 | regulation of protein localization to cell surface(GO:2000008) negative regulation of protein localization to cell surface(GO:2000009) |
| 0.0 | 1.3 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.0 | 2.0 | GO:0009247 | glycolipid biosynthetic process(GO:0009247) |
| 0.0 | 1.3 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.0 | 0.7 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 3.2 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.2 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.0 | 1.8 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 6.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.2 | 0.7 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 1.0 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 1.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.3 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 2.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.2 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.2 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.6 | GO:0036122 | BMP binding(GO:0036122) |
| 0.2 | 5.0 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.2 | 1.3 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.2 | 0.7 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 1.2 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.1 | 2.0 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 0.4 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.7 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.1 | 1.2 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 0.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 2.0 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 1.6 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 3.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 1.1 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.6 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 2.0 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.3 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.7 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.7 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 1.0 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 1.2 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |