Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
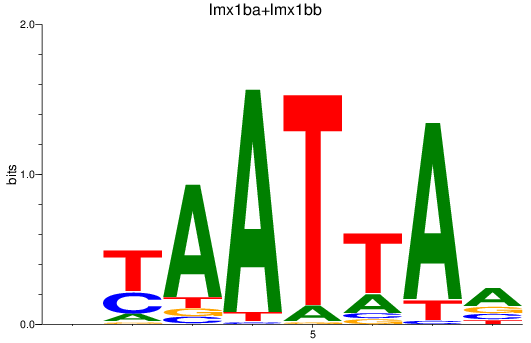
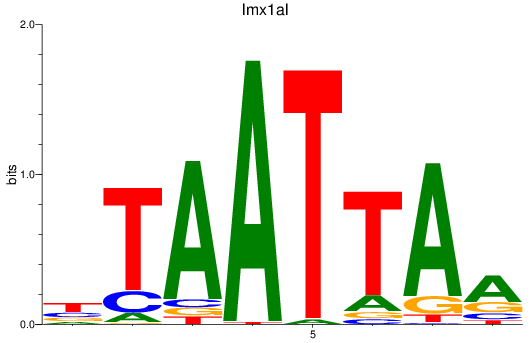
Results for lmx1ba+lmx1bb_lmx1al
Z-value: 0.68
Transcription factors associated with lmx1ba+lmx1bb_lmx1al
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
lmx1bb
|
ENSDARG00000068365 | LIM homeobox transcription factor 1, beta b |
|
lmx1ba
|
ENSDARG00000104815 | LIM homeobox transcription factor 1, beta a |
|
lmx1al
|
ENSDARG00000077915 | LIM homeobox transcription factor 1, alpha-like |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| lmx1bb | dr11_v1_chr8_-_33381913_33381913 | -0.31 | 4.2e-01 | Click! |
| lmx1ba | dr11_v1_chr5_-_5147041_5147041 | -0.28 | 4.6e-01 | Click! |
| lmx1al | dr11_v1_chr3_+_52806347_52806441 | -0.19 | 6.3e-01 | Click! |
Activity profile of lmx1ba+lmx1bb_lmx1al motif
Sorted Z-values of lmx1ba+lmx1bb_lmx1al motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_35830625 | 1.52 |
ENSDART00000180028
|
CU459056.1
|
|
| chr16_+_23961276 | 1.07 |
ENSDART00000192754
|
apoeb
|
apolipoprotein Eb |
| chr11_-_45138857 | 1.04 |
ENSDART00000166501
|
cant1b
|
calcium activated nucleotidase 1b |
| chr18_+_24921587 | 0.98 |
ENSDART00000191345
|
rgma
|
repulsive guidance molecule family member a |
| chr2_+_33326522 | 0.89 |
ENSDART00000056655
|
klf17
|
Kruppel-like factor 17 |
| chr11_-_41853874 | 0.89 |
ENSDART00000002556
|
mrto4
|
MRT4 homolog, ribosome maturation factor |
| chr3_+_46635527 | 0.87 |
ENSDART00000153971
|
si:dkey-248g21.1
|
si:dkey-248g21.1 |
| chr14_+_45406299 | 0.83 |
ENSDART00000173142
ENSDART00000112377 |
map1lc3cl
|
microtubule-associated protein 1 light chain 3 gamma, like |
| chr24_+_9412450 | 0.81 |
ENSDART00000132724
|
si:ch211-285f17.1
|
si:ch211-285f17.1 |
| chr18_-_33979693 | 0.78 |
ENSDART00000021215
|
si:ch211-203b20.7
|
si:ch211-203b20.7 |
| chr16_+_23431189 | 0.78 |
ENSDART00000004679
|
icn
|
ictacalcin |
| chr24_-_21923930 | 0.74 |
ENSDART00000131944
|
tagln3b
|
transgelin 3b |
| chr17_+_30369396 | 0.64 |
ENSDART00000076611
|
greb1
|
growth regulation by estrogen in breast cancer 1 |
| chr11_+_35050253 | 0.64 |
ENSDART00000124800
|
fam212aa
|
family with sequence similarity 212, member Aa |
| chr4_+_306036 | 0.64 |
ENSDART00000103659
|
msgn1
|
mesogenin 1 |
| chr24_-_6078222 | 0.61 |
ENSDART00000146830
|
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr1_-_5455498 | 0.60 |
ENSDART00000040368
ENSDART00000114035 |
mnx2b
|
motor neuron and pancreas homeobox 2b |
| chr16_+_33902006 | 0.57 |
ENSDART00000161807
ENSDART00000159474 |
gnl2
|
guanine nucleotide binding protein-like 2 (nucleolar) |
| chr13_+_27232848 | 0.56 |
ENSDART00000138043
|
rin2
|
Ras and Rab interactor 2 |
| chr7_+_56577906 | 0.55 |
ENSDART00000184023
|
hp
|
haptoglobin |
| chr7_+_19552381 | 0.55 |
ENSDART00000169060
|
si:ch211-212k18.5
|
si:ch211-212k18.5 |
| chr18_-_19456269 | 0.53 |
ENSDART00000060363
|
rpl4
|
ribosomal protein L4 |
| chr3_+_27798094 | 0.51 |
ENSDART00000075100
ENSDART00000151437 |
carhsp1
|
calcium regulated heat stable protein 1 |
| chr6_+_41191482 | 0.50 |
ENSDART00000000877
|
opn1mw3
|
opsin 1 (cone pigments), medium-wave-sensitive, 3 |
| chr7_+_20503344 | 0.49 |
ENSDART00000157699
|
si:dkey-19b23.12
|
si:dkey-19b23.12 |
| chr1_+_52392511 | 0.47 |
ENSDART00000144025
|
si:ch211-217k17.8
|
si:ch211-217k17.8 |
| chr2_-_28085884 | 0.45 |
ENSDART00000131506
|
cdh6
|
cadherin 6 |
| chr23_-_17003533 | 0.44 |
ENSDART00000080545
|
dnmt3bb.2
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate b.2 |
| chr15_-_21877726 | 0.44 |
ENSDART00000127819
ENSDART00000145646 ENSDART00000100897 ENSDART00000144739 |
zgc:162608
|
zgc:162608 |
| chr25_+_13620555 | 0.44 |
ENSDART00000163642
|
si:ch211-172l8.4
|
si:ch211-172l8.4 |
| chr18_-_14677936 | 0.44 |
ENSDART00000111995
|
si:dkey-238o13.4
|
si:dkey-238o13.4 |
| chr10_-_15340362 | 0.42 |
ENSDART00000148119
ENSDART00000127277 ENSDART00000154037 ENSDART00000189109 |
pum3
|
pumilio RNA-binding family member 3 |
| chr25_-_20378721 | 0.42 |
ENSDART00000181707
|
kctd15a
|
potassium channel tetramerization domain containing 15a |
| chr9_+_25776971 | 0.42 |
ENSDART00000146011
|
zeb2a
|
zinc finger E-box binding homeobox 2a |
| chr4_-_2545310 | 0.42 |
ENSDART00000150619
ENSDART00000140760 |
e2f7
|
E2F transcription factor 7 |
| chr2_-_7246848 | 0.42 |
ENSDART00000146434
|
zgc:153115
|
zgc:153115 |
| chr13_+_22295905 | 0.42 |
ENSDART00000180133
ENSDART00000181125 |
usp54a
|
ubiquitin specific peptidase 54a |
| chr3_+_23092762 | 0.40 |
ENSDART00000142884
ENSDART00000024136 |
gngt2a
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2a |
| chr1_-_10821584 | 0.40 |
ENSDART00000167452
ENSDART00000162137 |
crfb15
|
cytokine receptor family member B15 |
| chr16_+_29509133 | 0.39 |
ENSDART00000112116
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr16_+_26777473 | 0.39 |
ENSDART00000188870
|
cdh17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr9_-_20372977 | 0.39 |
ENSDART00000113418
|
igsf3
|
immunoglobulin superfamily, member 3 |
| chr24_-_4450238 | 0.38 |
ENSDART00000066835
|
fzd8a
|
frizzled class receptor 8a |
| chr10_-_43771447 | 0.38 |
ENSDART00000052307
|
arrdc3b
|
arrestin domain containing 3b |
| chr17_-_48944465 | 0.38 |
ENSDART00000154110
|
si:ch1073-80i24.3
|
si:ch1073-80i24.3 |
| chr12_+_10631266 | 0.38 |
ENSDART00000161455
|
csf3a
|
colony stimulating factor 3 (granulocyte) a |
| chr8_+_21353878 | 0.37 |
ENSDART00000056420
|
alas2
|
aminolevulinate, delta-, synthase 2 |
| chr15_-_22074315 | 0.37 |
ENSDART00000149830
|
drd2a
|
dopamine receptor D2a |
| chr17_-_26867725 | 0.37 |
ENSDART00000153590
|
si:dkey-221l4.10
|
si:dkey-221l4.10 |
| chr18_+_3037998 | 0.37 |
ENSDART00000185844
ENSDART00000162657 |
rps3
|
ribosomal protein S3 |
| chr4_+_16885854 | 0.35 |
ENSDART00000017726
|
etnk1
|
ethanolamine kinase 1 |
| chr7_+_56577522 | 0.35 |
ENSDART00000149130
ENSDART00000149624 |
hp
|
haptoglobin |
| chr1_+_45056371 | 0.34 |
ENSDART00000073689
ENSDART00000167309 |
btr01
|
bloodthirsty-related gene family, member 1 |
| chr2_-_30668580 | 0.33 |
ENSDART00000087270
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr9_-_12424791 | 0.33 |
ENSDART00000135447
ENSDART00000088199 |
zgc:162707
|
zgc:162707 |
| chr10_+_16092671 | 0.33 |
ENSDART00000182761
ENSDART00000154835 |
megf10
|
multiple EGF-like-domains 10 |
| chr6_+_39098397 | 0.32 |
ENSDART00000003716
ENSDART00000188655 |
prss60.2
|
protease, serine, 60.2 |
| chr15_+_34988148 | 0.32 |
ENSDART00000076269
|
ccdc105
|
coiled-coil domain containing 105 |
| chr4_+_72723304 | 0.32 |
ENSDART00000186791
ENSDART00000158902 ENSDART00000191925 |
rab3ip
|
RAB3A interacting protein (rabin3) |
| chr10_+_42423318 | 0.32 |
ENSDART00000134282
|
npy8ar
|
neuropeptide Y receptor Y8a |
| chr20_+_52554352 | 0.31 |
ENSDART00000153217
ENSDART00000145230 |
eef1db
|
eukaryotic translation elongation factor 1 delta b (guanine nucleotide exchange protein) |
| chr5_-_41307550 | 0.30 |
ENSDART00000143446
|
npr3
|
natriuretic peptide receptor 3 |
| chr18_+_38807239 | 0.30 |
ENSDART00000184332
|
fam214a
|
family with sequence similarity 214, member A |
| chr8_+_6576940 | 0.29 |
ENSDART00000138135
|
vsig8b
|
V-set and immunoglobulin domain containing 8b |
| chr17_-_42492668 | 0.29 |
ENSDART00000183946
|
CR925755.2
|
|
| chr23_-_24394719 | 0.28 |
ENSDART00000044918
|
epha2b
|
eph receptor A2 b |
| chr23_+_31913292 | 0.28 |
ENSDART00000136910
|
armc1l
|
armadillo repeat containing 1, like |
| chr10_+_7593185 | 0.28 |
ENSDART00000162617
ENSDART00000162590 ENSDART00000171744 |
ppp2cb
|
protein phosphatase 2, catalytic subunit, beta isozyme |
| chr8_-_23612462 | 0.27 |
ENSDART00000025024
|
slc38a5b
|
solute carrier family 38, member 5b |
| chr22_+_16535575 | 0.27 |
ENSDART00000083063
|
tal1
|
T-cell acute lymphocytic leukemia 1 |
| chr8_-_19216657 | 0.27 |
ENSDART00000135096
ENSDART00000135869 ENSDART00000145951 |
si:ch73-222f22.2
si:ch73-222f22.2
|
si:ch73-222f22.2 si:ch73-222f22.2 |
| chr1_+_52929185 | 0.27 |
ENSDART00000147683
|
inpp4b
|
inositol polyphosphate-4-phosphatase type II B |
| chr24_-_40860603 | 0.27 |
ENSDART00000188032
|
CU633479.7
|
|
| chr17_+_24722646 | 0.27 |
ENSDART00000138356
|
mtfr1l
|
mitochondrial fission regulator 1-like |
| chr2_+_20793982 | 0.26 |
ENSDART00000014785
|
prg4a
|
proteoglycan 4a |
| chr9_-_1702648 | 0.26 |
ENSDART00000102934
|
hnrnpa3
|
heterogeneous nuclear ribonucleoprotein A3 |
| chr19_-_11106315 | 0.26 |
ENSDART00000059102
|
arhgef1a
|
Rho guanine nucleotide exchange factor (GEF) 1a |
| chr3_-_23406964 | 0.26 |
ENSDART00000114723
|
rapgefl1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr22_+_2937485 | 0.26 |
ENSDART00000082222
ENSDART00000135507 ENSDART00000143258 |
cep19
|
centrosomal protein 19 |
| chr14_+_33329420 | 0.25 |
ENSDART00000171090
ENSDART00000164062 |
sowahd
|
sosondowah ankyrin repeat domain family d |
| chr7_+_42461850 | 0.25 |
ENSDART00000190350
|
adamts18
|
ADAM metallopeptidase with thrombospondin type 1 motif, 18 |
| chr5_-_20194876 | 0.25 |
ENSDART00000122587
|
dao.1
|
D-amino-acid oxidase, tandem duplicate 1 |
| chr4_+_31405646 | 0.25 |
ENSDART00000128732
|
si:rp71-5o12.3
|
si:rp71-5o12.3 |
| chr3_+_1637358 | 0.25 |
ENSDART00000180266
|
CR394546.5
|
|
| chr6_+_45918981 | 0.25 |
ENSDART00000149642
|
h6pd
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr20_-_31075972 | 0.24 |
ENSDART00000122927
|
si:ch211-198b3.4
|
si:ch211-198b3.4 |
| chr22_+_1291651 | 0.24 |
ENSDART00000159296
|
si:ch73-138e16.4
|
si:ch73-138e16.4 |
| chr7_-_30174882 | 0.24 |
ENSDART00000110409
|
frmd5
|
FERM domain containing 5 |
| chr22_-_6988102 | 0.24 |
ENSDART00000185618
|
fgfr1bl
|
fibroblast growth factor receptor 1b, like |
| chr4_+_12612145 | 0.24 |
ENSDART00000181201
|
lmo3
|
LIM domain only 3 |
| chr23_+_11285662 | 0.24 |
ENSDART00000111028
|
chl1a
|
cell adhesion molecule L1-like a |
| chr15_-_43284021 | 0.24 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr7_+_20471315 | 0.24 |
ENSDART00000173714
|
si:dkey-19b23.13
|
si:dkey-19b23.13 |
| chr16_-_51299061 | 0.24 |
ENSDART00000148677
|
serpinb1l4
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1, like 4 |
| chr15_-_2493771 | 0.23 |
ENSDART00000184906
|
neu4
|
sialidase 4 |
| chr16_-_29528198 | 0.23 |
ENSDART00000150028
|
onecutl
|
one cut domain, family member, like |
| chr10_+_11767791 | 0.23 |
ENSDART00000092047
|
ppwd1
|
peptidylprolyl isomerase domain and WD repeat containing 1 |
| chr24_+_21540842 | 0.23 |
ENSDART00000091529
|
wasf3b
|
WAS protein family, member 3b |
| chr7_+_7696665 | 0.23 |
ENSDART00000091099
|
ino80b
|
INO80 complex subunit B |
| chr2_+_19522082 | 0.22 |
ENSDART00000146098
|
pimr49
|
Pim proto-oncogene, serine/threonine kinase, related 49 |
| chr22_-_12160283 | 0.22 |
ENSDART00000146785
ENSDART00000128176 |
tmem163b
|
transmembrane protein 163b |
| chr16_-_29387215 | 0.21 |
ENSDART00000148787
|
s100a1
|
S100 calcium binding protein A1 |
| chr16_-_42894628 | 0.21 |
ENSDART00000045600
|
hfe2
|
hemochromatosis type 2 |
| chr5_-_12219572 | 0.21 |
ENSDART00000167834
|
nos1
|
nitric oxide synthase 1 (neuronal) |
| chr17_+_23298928 | 0.21 |
ENSDART00000153652
|
zgc:165461
|
zgc:165461 |
| chr20_-_22476255 | 0.21 |
ENSDART00000103510
|
pdgfra
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr7_-_28148310 | 0.21 |
ENSDART00000044208
|
lmo1
|
LIM domain only 1 |
| chr3_-_39695856 | 0.21 |
ENSDART00000148247
|
b9d1
|
B9 protein domain 1 |
| chr8_+_23152244 | 0.21 |
ENSDART00000036801
ENSDART00000184298 |
slc17a9a
|
solute carrier family 17 (vesicular nucleotide transporter), member 9a |
| chr7_+_72003301 | 0.21 |
ENSDART00000012918
ENSDART00000182268 ENSDART00000185750 |
psmd9
|
proteasome 26S subunit, non-ATPase 9 |
| chr15_+_857148 | 0.21 |
ENSDART00000156949
|
si:dkey-7i4.13
|
si:dkey-7i4.13 |
| chr20_+_11731039 | 0.21 |
ENSDART00000152215
ENSDART00000152585 |
si:ch211-155o21.3
|
si:ch211-155o21.3 |
| chr22_+_38037530 | 0.21 |
ENSDART00000012212
|
commd2
|
COMM domain containing 2 |
| chr19_+_43119014 | 0.20 |
ENSDART00000023156
|
eef1a1l1
|
eukaryotic translation elongation factor 1 alpha 1, like 1 |
| chr23_-_31974060 | 0.20 |
ENSDART00000168087
|
BX927210.1
|
|
| chr20_-_37490612 | 0.20 |
ENSDART00000185458
|
si:ch211-202p1.5
|
si:ch211-202p1.5 |
| chr16_+_23087326 | 0.20 |
ENSDART00000167518
ENSDART00000161087 |
efna3b
|
ephrin-A3b |
| chr23_-_16485190 | 0.20 |
ENSDART00000155038
|
si:dkeyp-100a5.4
|
si:dkeyp-100a5.4 |
| chr16_+_13883872 | 0.19 |
ENSDART00000101304
ENSDART00000136005 |
atg12
|
ATG12 autophagy related 12 homolog (S. cerevisiae) |
| chr10_+_16584382 | 0.19 |
ENSDART00000112039
|
CR790388.1
|
|
| chr10_+_21867307 | 0.19 |
ENSDART00000126629
|
cbln17
|
cerebellin 17 |
| chr4_-_20292821 | 0.19 |
ENSDART00000136069
ENSDART00000192504 |
cacna2d4a
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4a |
| chr3_+_17537352 | 0.18 |
ENSDART00000104549
|
hcrt
|
hypocretin (orexin) neuropeptide precursor |
| chr22_-_2937503 | 0.18 |
ENSDART00000092991
ENSDART00000131110 |
pigx
|
phosphatidylinositol glycan anchor biosynthesis, class X |
| chr24_-_30862168 | 0.18 |
ENSDART00000168540
|
ptbp2a
|
polypyrimidine tract binding protein 2a |
| chr8_-_12867434 | 0.18 |
ENSDART00000081657
|
slc2a6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr20_+_539852 | 0.17 |
ENSDART00000185994
|
dse
|
dermatan sulfate epimerase |
| chr15_-_4528326 | 0.17 |
ENSDART00000158122
ENSDART00000155619 ENSDART00000128602 |
tfdp2
|
transcription factor Dp-2 |
| chr16_+_23913943 | 0.17 |
ENSDART00000175404
ENSDART00000129525 |
apoa4b.1
|
apolipoprotein A-IV b, tandem duplicate 1 |
| chr17_-_23416897 | 0.17 |
ENSDART00000163391
|
si:ch211-149k12.3
|
si:ch211-149k12.3 |
| chr23_-_6749820 | 0.17 |
ENSDART00000128772
|
ptges3b
|
prostaglandin E synthase 3b (cytosolic) |
| chr25_+_3058924 | 0.17 |
ENSDART00000029580
|
fth1b
|
ferritin, heavy polypeptide 1b |
| chr15_+_5360407 | 0.17 |
ENSDART00000110420
|
or112-1
|
odorant receptor, family A, subfamily 112, member 1 |
| chr2_+_19633493 | 0.16 |
ENSDART00000147989
|
pimr54
|
Pim proto-oncogene, serine/threonine kinase, related 54 |
| chr8_-_37249813 | 0.16 |
ENSDART00000098634
ENSDART00000140233 ENSDART00000061328 |
rbm39b
|
RNA binding motif protein 39b |
| chr11_-_40728380 | 0.16 |
ENSDART00000023745
|
ccdc114
|
coiled-coil domain containing 114 |
| chr7_+_7048245 | 0.16 |
ENSDART00000001649
|
actn3b
|
actinin alpha 3b |
| chr20_-_52902693 | 0.16 |
ENSDART00000166115
ENSDART00000161050 |
ctsbb
|
cathepsin Bb |
| chr2_+_19578446 | 0.16 |
ENSDART00000164758
|
pimr50
|
Pim proto-oncogene, serine/threonine kinase, related 50 |
| chr21_-_40676224 | 0.16 |
ENSDART00000162623
|
arxb
|
aristaless related homeobox b |
| chr22_-_8725768 | 0.15 |
ENSDART00000189873
ENSDART00000181819 |
si:ch73-27e22.1
si:ch73-27e22.8
|
si:ch73-27e22.1 si:ch73-27e22.8 |
| chr24_-_35561672 | 0.15 |
ENSDART00000058564
|
mcm4
|
minichromosome maintenance complex component 4 |
| chr24_+_16985181 | 0.15 |
ENSDART00000135580
|
eif2s3
|
eukaryotic translation initiation factor 2, subunit 3 gamma |
| chr9_-_41077934 | 0.15 |
ENSDART00000100342
|
ankar
|
ankyrin and armadillo repeat containing |
| chr25_+_7671640 | 0.14 |
ENSDART00000145367
|
kcnj11l
|
potassium inwardly-rectifying channel, subfamily J, member 11, like |
| chr4_+_22480169 | 0.14 |
ENSDART00000146272
ENSDART00000066904 |
ndufb2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2 |
| chr23_+_19813677 | 0.14 |
ENSDART00000139192
ENSDART00000142308 |
emd
|
emerin (Emery-Dreifuss muscular dystrophy) |
| chr20_-_43663494 | 0.14 |
ENSDART00000144564
|
BX470188.1
|
|
| chr11_+_18873113 | 0.14 |
ENSDART00000103969
ENSDART00000103968 |
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr5_+_42400777 | 0.14 |
ENSDART00000183114
|
BX548073.8
|
|
| chr1_+_33668236 | 0.14 |
ENSDART00000122316
ENSDART00000102184 |
arl13b
|
ADP-ribosylation factor-like 13b |
| chr1_+_12767318 | 0.14 |
ENSDART00000162652
|
pcdh10a
|
protocadherin 10a |
| chr1_+_34696503 | 0.14 |
ENSDART00000186106
|
CR339054.2
|
|
| chr19_+_45962016 | 0.14 |
ENSDART00000169710
|
utp23
|
UTP23, small subunit (SSU) processome component, homolog (yeast) |
| chr2_+_36721357 | 0.13 |
ENSDART00000019063
|
pdcd10b
|
programmed cell death 10b |
| chr3_-_39696066 | 0.13 |
ENSDART00000015393
|
b9d1
|
B9 protein domain 1 |
| chr24_-_31425799 | 0.13 |
ENSDART00000157998
|
cngb3.1
|
cyclic nucleotide gated channel beta 3, tandem duplicate 1 |
| chr13_+_25449681 | 0.13 |
ENSDART00000101328
|
atoh7
|
atonal bHLH transcription factor 7 |
| chr8_-_37249991 | 0.13 |
ENSDART00000189275
ENSDART00000178556 |
rbm39b
|
RNA binding motif protein 39b |
| chr10_+_11261576 | 0.13 |
ENSDART00000155333
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr3_-_21094437 | 0.13 |
ENSDART00000153739
ENSDART00000109790 |
nlk1
|
nemo-like kinase, type 1 |
| chr13_+_4225173 | 0.13 |
ENSDART00000058242
ENSDART00000143456 |
mea1
|
male-enhanced antigen 1 |
| chr14_+_34490445 | 0.13 |
ENSDART00000132193
ENSDART00000148044 |
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr10_-_33297864 | 0.12 |
ENSDART00000163360
|
PRDM15
|
PR/SET domain 15 |
| chr5_-_42661012 | 0.12 |
ENSDART00000158339
|
pimr58
|
Pim proto-oncogene, serine/threonine kinase, related 58 |
| chr5_-_67750907 | 0.12 |
ENSDART00000172097
|
b4galt4
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 4 |
| chr13_+_45431660 | 0.12 |
ENSDART00000099950
|
syf2
|
SYF2 pre-mRNA-splicing factor |
| chr11_-_41220794 | 0.12 |
ENSDART00000192895
|
mrps16
|
mitochondrial ribosomal protein S16 |
| chr17_-_11439815 | 0.12 |
ENSDART00000130105
|
psma3
|
proteasome subunit alpha 3 |
| chr12_+_16168342 | 0.12 |
ENSDART00000079326
ENSDART00000170024 |
lrp2b
|
low density lipoprotein receptor-related protein 2b |
| chr4_-_43388943 | 0.12 |
ENSDART00000150796
|
si:dkey-29j8.2
|
si:dkey-29j8.2 |
| chr18_+_19456648 | 0.12 |
ENSDART00000079695
|
zwilch
|
zwilch kinetochore protein |
| chr16_-_55028740 | 0.11 |
ENSDART00000156368
ENSDART00000161704 |
zgc:114181
|
zgc:114181 |
| chr17_+_10593398 | 0.11 |
ENSDART00000168897
ENSDART00000193989 ENSDART00000191664 ENSDART00000167188 |
mapkbp1
|
mitogen-activated protein kinase binding protein 1 |
| chr17_+_23462972 | 0.11 |
ENSDART00000112959
ENSDART00000192168 |
ankrd1a
|
ankyrin repeat domain 1a (cardiac muscle) |
| chr4_+_34657691 | 0.11 |
ENSDART00000158655
|
AL645691.1
|
|
| chr4_-_1801519 | 0.11 |
ENSDART00000188604
ENSDART00000135749 |
nudt4b
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4b |
| chr6_+_52350443 | 0.11 |
ENSDART00000151612
ENSDART00000151349 |
si:ch211-239j9.1
|
si:ch211-239j9.1 |
| chr14_+_30795559 | 0.11 |
ENSDART00000006132
|
cfl1
|
cofilin 1 |
| chr17_-_10122204 | 0.11 |
ENSDART00000160751
|
BX088587.1
|
|
| chr2_+_29842878 | 0.11 |
ENSDART00000131376
|
si:ch211-207d6.2
|
si:ch211-207d6.2 |
| chr24_-_17067284 | 0.11 |
ENSDART00000111237
|
armc3
|
armadillo repeat containing 3 |
| chr3_-_58798815 | 0.11 |
ENSDART00000082920
|
si:ch73-281f12.4
|
si:ch73-281f12.4 |
| chr4_-_9891874 | 0.11 |
ENSDART00000067193
|
adm2a
|
adrenomedullin 2a |
| chr17_+_21964472 | 0.11 |
ENSDART00000063704
ENSDART00000188904 |
crip3
|
cysteine-rich protein 3 |
| chr13_+_35339182 | 0.11 |
ENSDART00000019323
|
jag1b
|
jagged 1b |
| chr13_-_12602920 | 0.11 |
ENSDART00000102311
|
lrit3b
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 3b |
| chr8_+_16758304 | 0.11 |
ENSDART00000133514
|
elovl7a
|
ELOVL fatty acid elongase 7a |
| chr18_-_2433011 | 0.11 |
ENSDART00000181922
ENSDART00000193276 |
CR769778.1
|
|
| chr2_+_43469241 | 0.11 |
ENSDART00000142078
ENSDART00000098265 |
nrp1b
|
neuropilin 1b |
| chr14_-_22113600 | 0.11 |
ENSDART00000113752
|
si:dkey-6i22.5
|
si:dkey-6i22.5 |
| chr21_-_23331619 | 0.11 |
ENSDART00000007806
|
zbtb16a
|
zinc finger and BTB domain containing 16a |
| chr2_+_24507597 | 0.11 |
ENSDART00000133109
|
rps28
|
ribosomal protein S28 |
| chr22_-_621609 | 0.11 |
ENSDART00000137264
ENSDART00000106636 |
srsf3b
|
serine/arginine-rich splicing factor 3b |
| chr1_-_46875493 | 0.11 |
ENSDART00000115081
|
agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr19_-_5699703 | 0.11 |
ENSDART00000082050
|
zgc:174904
|
zgc:174904 |
| chr15_+_1796313 | 0.10 |
ENSDART00000126253
|
fam124b
|
family with sequence similarity 124B |
| chr4_+_12612723 | 0.10 |
ENSDART00000133767
|
lmo3
|
LIM domain only 3 |
| chr8_+_23355484 | 0.10 |
ENSDART00000085361
ENSDART00000125729 |
dnmt3ba
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate a |
| chr4_-_16334362 | 0.10 |
ENSDART00000101461
|
epyc
|
epiphycan |
Network of associatons between targets according to the STRING database.
First level regulatory network of lmx1ba+lmx1bb_lmx1al
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0071830 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 0.3 | 0.9 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.2 | 0.6 | GO:0048341 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) |
| 0.2 | 1.0 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 0.6 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.1 | 0.4 | GO:0060898 | spinal cord radial glial cell differentiation(GO:0021531) eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.1 | 0.3 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.1 | 0.4 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.1 | 0.4 | GO:0034380 | regulation of cholesterol esterification(GO:0010872) positive regulation of cholesterol esterification(GO:0010873) high-density lipoprotein particle assembly(GO:0034380) |
| 0.1 | 0.4 | GO:0032875 | regulation of DNA endoreduplication(GO:0032875) positive regulation of DNA endoreduplication(GO:0032877) DNA endoreduplication(GO:0042023) |
| 0.1 | 0.2 | GO:0039015 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.1 | 0.2 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) retrograde trans-synaptic signaling by nitric oxide(GO:0098924) |
| 0.1 | 0.3 | GO:0089709 | L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.1 | 0.3 | GO:0046436 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.1 | 0.4 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.1 | 0.3 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.1 | GO:1903441 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to ciliary membrane(GO:1903441) |
| 0.0 | 0.2 | GO:0042755 | eating behavior(GO:0042755) |
| 0.0 | 0.1 | GO:1903358 | regulation of Golgi organization(GO:1903358) |
| 0.0 | 0.6 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.2 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.0 | 0.2 | GO:0003100 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.2 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.0 | 0.1 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.4 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.9 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.2 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.0 | 0.1 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.4 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.2 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.6 | GO:0021983 | pituitary gland development(GO:0021983) |
| 0.0 | 0.4 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.0 | 0.1 | GO:0015988 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) electron transport coupled proton transport(GO:0015990) |
| 0.0 | 0.2 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.1 | GO:1901906 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.1 | GO:0010660 | negative regulation of muscle cell apoptotic process(GO:0010656) muscle cell apoptotic process(GO:0010657) striated muscle cell apoptotic process(GO:0010658) regulation of muscle cell apoptotic process(GO:0010660) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 0.0 | 0.4 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.4 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.1 | GO:0097435 | fibril organization(GO:0097435) |
| 0.0 | 0.2 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.0 | 0.2 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.4 | GO:0007622 | rhythmic behavior(GO:0007622) circadian behavior(GO:0048512) |
| 0.0 | 0.4 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.0 | 0.3 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 0.1 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.4 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 0.1 | GO:1902946 | positive regulation of receptor-mediated endocytosis(GO:0048260) protein localization to early endosome(GO:1902946) |
| 0.0 | 0.2 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.1 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.0 | 0.3 | GO:0060038 | cardiac muscle cell proliferation(GO:0060038) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.1 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.0 | 0.2 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.1 | GO:0072395 | signal transduction involved in cell cycle checkpoint(GO:0072395) |
| 0.0 | 0.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.0 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.0 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.0 | 0.3 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) dendritic spine organization(GO:0097061) |
| 0.0 | 0.4 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.0 | GO:1900364 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.1 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.0 | 0.0 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.0 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.1 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.1 | 0.4 | GO:0043220 | compact myelin(GO:0043218) Schmidt-Lanterman incisure(GO:0043220) |
| 0.1 | 0.3 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.1 | 0.2 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.0 | 0.1 | GO:0090443 | FAR/SIN/STRIPAK complex(GO:0090443) |
| 0.0 | 0.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 1.0 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.4 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.9 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.1 | GO:0098556 | cytoplasmic side of rough endoplasmic reticulum membrane(GO:0098556) |
| 0.0 | 0.2 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.1 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.0 | 0.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.4 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.2 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.1 | GO:0071540 | eukaryotic translation initiation factor 3 complex, eIF3e(GO:0071540) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.2 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.8 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.0 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 0.4 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.1 | 0.4 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.1 | 0.2 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.1 | 0.3 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.1 | 0.3 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.1 | 0.4 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.1 | 0.2 | GO:0048407 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 1.0 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.2 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.5 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.2 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.9 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 1.0 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.2 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.1 | GO:0047173 | phosphatidylcholine-retinol O-acyltransferase activity(GO:0047173) |
| 0.0 | 0.2 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 0.0 | 0.2 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.2 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 0.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.1 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.0 | 0.4 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.4 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.1 | GO:0017064 | fatty acid amide hydrolase activity(GO:0017064) |
| 0.0 | 0.2 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.3 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:0034432 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.2 | GO:0052796 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.1 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.1 | GO:0072545 | tyrosine binding(GO:0072545) |
| 0.0 | 0.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.4 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.0 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.0 | 0.2 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.1 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.0 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.1 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 1.1 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.5 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.1 | GO:0031843 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.5 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 1.3 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.3 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.6 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.2 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.1 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |