Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
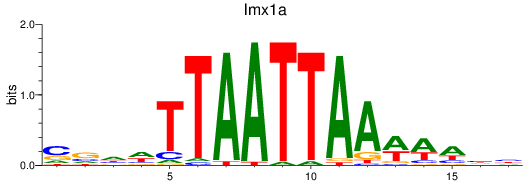
Results for lmx1a
Z-value: 0.73
Transcription factors associated with lmx1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
lmx1a
|
ENSDARG00000020354 | LIM homeobox transcription factor 1, alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| lmx1a | dr11_v1_chr20_+_33922339_33922339 | -0.87 | 2.0e-03 | Click! |
Activity profile of lmx1a motif
Sorted Z-values of lmx1a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr24_-_26328721 | 1.59 |
ENSDART00000125468
|
apodb
|
apolipoprotein Db |
| chr12_+_25600685 | 1.48 |
ENSDART00000077157
|
six3b
|
SIX homeobox 3b |
| chr24_+_24461558 | 1.16 |
ENSDART00000182424
|
bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr16_-_16152199 | 1.13 |
ENSDART00000012718
|
fabp11b
|
fatty acid binding protein 11b |
| chr24_+_24461341 | 1.13 |
ENSDART00000147658
|
bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr7_+_7048245 | 1.08 |
ENSDART00000001649
|
actn3b
|
actinin alpha 3b |
| chr9_+_34641237 | 1.02 |
ENSDART00000133996
|
shox
|
short stature homeobox |
| chr16_-_28856112 | 0.99 |
ENSDART00000078543
|
syt11b
|
synaptotagmin XIb |
| chr12_-_35830625 | 0.99 |
ENSDART00000180028
|
CU459056.1
|
|
| chr6_+_9130989 | 0.96 |
ENSDART00000162588
|
rgn
|
regucalcin |
| chr2_-_30668580 | 0.96 |
ENSDART00000087270
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr22_-_24285432 | 0.89 |
ENSDART00000164083
|
si:ch211-117l17.4
|
si:ch211-117l17.4 |
| chr2_+_2223837 | 0.86 |
ENSDART00000101038
ENSDART00000129354 |
tmie
|
transmembrane inner ear |
| chr7_-_58729894 | 0.86 |
ENSDART00000149347
|
chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr8_-_14484599 | 0.85 |
ENSDART00000057644
|
lhx4
|
LIM homeobox 4 |
| chr24_-_38657683 | 0.85 |
ENSDART00000154843
|
si:ch1073-164k15.3
|
si:ch1073-164k15.3 |
| chr18_+_46382484 | 0.82 |
ENSDART00000024202
ENSDART00000142790 |
daw1
|
dynein assembly factor with WDR repeat domains 1 |
| chr16_-_12173554 | 0.81 |
ENSDART00000110567
ENSDART00000155935 |
clstn3
|
calsyntenin 3 |
| chr1_-_669717 | 0.76 |
ENSDART00000160564
|
cyyr1
|
cysteine/tyrosine-rich 1 |
| chr7_-_38658411 | 0.75 |
ENSDART00000109463
ENSDART00000017155 |
npsn
|
nephrosin |
| chr4_+_72723304 | 0.74 |
ENSDART00000186791
ENSDART00000158902 ENSDART00000191925 |
rab3ip
|
RAB3A interacting protein (rabin3) |
| chr20_-_45812144 | 0.73 |
ENSDART00000147897
ENSDART00000147637 |
fermt1
|
fermitin family member 1 |
| chr12_+_47122104 | 0.70 |
ENSDART00000184248
|
CABZ01088982.1
|
|
| chr15_-_22074315 | 0.70 |
ENSDART00000149830
|
drd2a
|
dopamine receptor D2a |
| chr5_-_31901468 | 0.70 |
ENSDART00000147814
ENSDART00000141446 |
coro1cb
|
coronin, actin binding protein, 1Cb |
| chr15_+_9861973 | 0.69 |
ENSDART00000170945
|
si:dkey-13m3.2
|
si:dkey-13m3.2 |
| chr15_+_47440477 | 0.67 |
ENSDART00000002384
|
phox2a
|
paired-like homeobox 2a |
| chr23_-_39666519 | 0.65 |
ENSDART00000110868
ENSDART00000190961 |
vwa1
|
von Willebrand factor A domain containing 1 |
| chr8_-_12867434 | 0.60 |
ENSDART00000081657
|
slc2a6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr19_+_14573998 | 0.60 |
ENSDART00000022076
|
fam46bb
|
family with sequence similarity 46, member Bb |
| chr9_+_37152564 | 0.58 |
ENSDART00000189497
|
gli2a
|
GLI family zinc finger 2a |
| chr22_-_12160283 | 0.57 |
ENSDART00000146785
ENSDART00000128176 |
tmem163b
|
transmembrane protein 163b |
| chr1_+_6172786 | 0.57 |
ENSDART00000126468
|
prkag3a
|
protein kinase, AMP-activated, gamma 3a non-catalytic subunit |
| chr3_+_50312422 | 0.53 |
ENSDART00000157689
|
gas7a
|
growth arrest-specific 7a |
| chr24_-_40860603 | 0.52 |
ENSDART00000188032
|
CU633479.7
|
|
| chr16_-_12173399 | 0.49 |
ENSDART00000142574
|
clstn3
|
calsyntenin 3 |
| chr3_+_14259115 | 0.49 |
ENSDART00000154470
|
si:ch211-108d22.1
|
si:ch211-108d22.1 |
| chr8_+_43340995 | 0.48 |
ENSDART00000038566
|
rflna
|
refilin A |
| chr14_-_451555 | 0.45 |
ENSDART00000190906
|
FAT4
|
FAT atypical cadherin 4 |
| chr2_+_36701322 | 0.42 |
ENSDART00000002510
|
golim4b
|
golgi integral membrane protein 4b |
| chr24_+_21540842 | 0.42 |
ENSDART00000091529
|
wasf3b
|
WAS protein family, member 3b |
| chr22_-_19552796 | 0.38 |
ENSDART00000148088
ENSDART00000105485 |
si:dkey-78l4.14
|
si:dkey-78l4.14 |
| chr20_+_52389858 | 0.37 |
ENSDART00000185863
ENSDART00000166651 |
arhgap39
|
Rho GTPase activating protein 39 |
| chr19_+_32158010 | 0.35 |
ENSDART00000005255
|
mrpl53
|
mitochondrial ribosomal protein L53 |
| chr9_+_24159280 | 0.35 |
ENSDART00000184624
ENSDART00000178422 |
hecw2a
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2a |
| chr13_-_29421331 | 0.33 |
ENSDART00000150228
|
chata
|
choline O-acetyltransferase a |
| chr4_+_57881965 | 0.32 |
ENSDART00000162234
|
si:dkeyp-44b5.4
|
si:dkeyp-44b5.4 |
| chr6_+_3004972 | 0.29 |
ENSDART00000186750
ENSDART00000183862 ENSDART00000191485 ENSDART00000171014 |
ptprfa
|
protein tyrosine phosphatase, receptor type, f, a |
| chr23_-_16485190 | 0.29 |
ENSDART00000155038
|
si:dkeyp-100a5.4
|
si:dkeyp-100a5.4 |
| chr18_-_17485419 | 0.27 |
ENSDART00000018764
|
foxl1
|
forkhead box L1 |
| chr8_-_12867128 | 0.26 |
ENSDART00000142201
|
slc2a6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr5_-_30715225 | 0.22 |
ENSDART00000016758
|
ftr82
|
finTRIM family, member 82 |
| chr10_+_16584382 | 0.22 |
ENSDART00000112039
|
CR790388.1
|
|
| chr17_-_23616626 | 0.19 |
ENSDART00000104730
|
ifit14
|
interferon-induced protein with tetratricopeptide repeats 14 |
| chr22_-_8725768 | 0.19 |
ENSDART00000189873
ENSDART00000181819 |
si:ch73-27e22.1
si:ch73-27e22.8
|
si:ch73-27e22.1 si:ch73-27e22.8 |
| chr8_-_30979494 | 0.18 |
ENSDART00000138959
|
si:ch211-251j10.3
|
si:ch211-251j10.3 |
| chr4_+_45484774 | 0.18 |
ENSDART00000150573
|
si:dkey-256i11.6
|
si:dkey-256i11.6 |
| chr11_-_2838699 | 0.18 |
ENSDART00000066189
|
lhfpl5a
|
LHFPL tetraspan subfamily member 5a |
| chr21_+_25802190 | 0.17 |
ENSDART00000128987
|
nf2b
|
neurofibromin 2b (merlin) |
| chr2_-_16217344 | 0.16 |
ENSDART00000152031
|
arhgef4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr1_+_51039558 | 0.16 |
ENSDART00000024743
|
dpy30
|
dpy-30 histone methyltransferase complex regulatory subunit |
| chr17_-_45040813 | 0.16 |
ENSDART00000075514
|
entpd5a
|
ectonucleoside triphosphate diphosphohydrolase 5a |
| chr25_+_16189044 | 0.15 |
ENSDART00000143975
|
si:dkey-80c24.1
|
si:dkey-80c24.1 |
| chr20_+_6035427 | 0.15 |
ENSDART00000054086
|
tshr
|
thyroid stimulating hormone receptor |
| chr25_-_16157776 | 0.12 |
ENSDART00000138453
|
si:dkey-80c24.5
|
si:dkey-80c24.5 |
| chr9_+_24159725 | 0.12 |
ENSDART00000137756
|
hecw2a
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2a |
| chr10_-_5844915 | 0.12 |
ENSDART00000185929
|
ankrd55
|
ankyrin repeat domain 55 |
| chr23_+_17878969 | 0.12 |
ENSDART00000154934
ENSDART00000154140 ENSDART00000156331 |
si:ch73-390p7.2
|
si:ch73-390p7.2 |
| chr20_-_9095105 | 0.10 |
ENSDART00000140792
|
oma1
|
OMA1 zinc metallopeptidase |
| chr22_+_19552987 | 0.09 |
ENSDART00000105315
|
hsd11b1la
|
hydroxysteroid (11-beta) dehydrogenase 1-like a |
| chr2_+_38373272 | 0.07 |
ENSDART00000113111
|
psmb5
|
proteasome subunit beta 5 |
| chr3_-_24093144 | 0.05 |
ENSDART00000103982
|
nfe2l1a
|
nuclear factor, erythroid 2-like 1a |
| chr19_+_46158078 | 0.04 |
ENSDART00000183933
ENSDART00000164055 |
cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr1_-_50438247 | 0.03 |
ENSDART00000114098
|
dkk2
|
dickkopf WNT signaling pathway inhibitor 2 |
| chr17_+_30369396 | 0.03 |
ENSDART00000076611
|
greb1
|
growth regulation by estrogen in breast cancer 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of lmx1a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0042364 | L-ascorbic acid metabolic process(GO:0019852) water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.2 | 2.1 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.2 | 0.5 | GO:1900157 | negative regulation of chondrocyte differentiation(GO:0032331) negative regulation of cartilage development(GO:0061037) negative regulation of chondrocyte development(GO:0061182) regulation of bone mineralization involved in bone maturation(GO:1900157) negative regulation of bone mineralization involved in bone maturation(GO:1900158) negative regulation of bone development(GO:1903011) |
| 0.1 | 1.3 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 0.3 | GO:1900620 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.1 | 0.9 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 | 0.2 | GO:0070589 | cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.1 | 0.7 | GO:0021703 | locus ceruleus development(GO:0021703) |
| 0.1 | 0.5 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.0 | 0.7 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 1.6 | GO:0007568 | aging(GO:0007568) |
| 0.0 | 0.9 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.8 | GO:0001539 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.0 | 0.3 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.0 | 1.0 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.4 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 1.2 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 0.6 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.6 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.2 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.6 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.5 | GO:0051904 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.0 | 0.1 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.0 | 0.1 | GO:0030816 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.0 | 0.2 | GO:0060122 | inner ear receptor stereocilium organization(GO:0060122) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.0 | 0.6 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.6 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.7 | GO:0098978 | glutamatergic synapse(GO:0098978) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 0.5 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.7 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.3 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 1.0 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.6 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 0.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.0 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |