Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
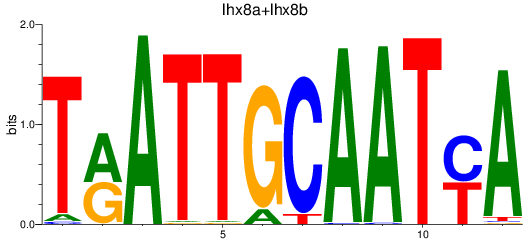
Results for lhx8a+lhx8b
Z-value: 1.16
Transcription factors associated with lhx8a+lhx8b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
lhx8a
|
ENSDARG00000002330 | LIM homeobox 8a |
|
lhx8b
|
ENSDARG00000042145 | LIM homeobox 8b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| lhx8b | dr11_v1_chr8_-_17926814_17926814 | 0.60 | 8.6e-02 | Click! |
| lhx8a | dr11_v1_chr2_+_11206317_11206317 | -0.26 | 5.0e-01 | Click! |
Activity profile of lhx8a+lhx8b motif
Sorted Z-values of lhx8a+lhx8b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_7546259 | 2.46 |
ENSDART00000015732
|
mylz3
|
myosin, light polypeptide 3, skeletal muscle |
| chr9_-_22318511 | 2.11 |
ENSDART00000129295
|
crygm2d2
|
crystallin, gamma M2d2 |
| chr25_+_31405266 | 1.93 |
ENSDART00000103395
|
tnnt3a
|
troponin T type 3a (skeletal, fast) |
| chr18_-_8877077 | 1.86 |
ENSDART00000137266
|
si:dkey-95h12.2
|
si:dkey-95h12.2 |
| chr9_-_22310919 | 1.78 |
ENSDART00000108719
|
crygm2d10
|
crystallin, gamma M2d10 |
| chr15_-_23645810 | 1.63 |
ENSDART00000168845
|
ckmb
|
creatine kinase, muscle b |
| chr5_-_46505691 | 1.56 |
ENSDART00000111589
ENSDART00000122966 ENSDART00000166907 |
hapln1a
|
hyaluronan and proteoglycan link protein 1a |
| chr10_+_21804772 | 1.54 |
ENSDART00000162194
|
pcdh1g31
|
protocadherin 1 gamma 31 |
| chr5_+_32247310 | 1.48 |
ENSDART00000182649
|
myha
|
myosin, heavy chain a |
| chr14_-_25577094 | 1.24 |
ENSDART00000163669
|
cplx2
|
complexin 2 |
| chr1_-_58064738 | 1.23 |
ENSDART00000073778
|
caspb
|
caspase b |
| chr12_+_42574148 | 1.18 |
ENSDART00000157855
|
ebf3a
|
early B cell factor 3a |
| chr12_-_4756478 | 1.11 |
ENSDART00000152181
|
mapta
|
microtubule-associated protein tau a |
| chr13_+_23988442 | 1.01 |
ENSDART00000010918
|
agt
|
angiotensinogen |
| chr3_+_25154078 | 0.99 |
ENSDART00000156973
|
si:ch211-256m1.8
|
si:ch211-256m1.8 |
| chr16_+_20926673 | 0.98 |
ENSDART00000009827
|
hoxa2b
|
homeobox A2b |
| chr21_+_13861589 | 0.96 |
ENSDART00000015629
ENSDART00000171306 |
stxbp1a
|
syntaxin binding protein 1a |
| chr6_-_58764672 | 0.96 |
ENSDART00000154322
|
soat2
|
sterol O-acyltransferase 2 |
| chr15_+_33991928 | 0.95 |
ENSDART00000170177
|
vwde
|
von Willebrand factor D and EGF domains |
| chr17_-_6730247 | 0.95 |
ENSDART00000031091
|
vsnl1b
|
visinin-like 1b |
| chr1_+_56274613 | 0.95 |
ENSDART00000052683
|
c3a.3
|
complement component c3a, duplicate 3 |
| chr6_+_40661703 | 0.94 |
ENSDART00000142492
|
eno1b
|
enolase 1b, (alpha) |
| chr10_-_43568239 | 0.90 |
ENSDART00000131731
ENSDART00000097433 ENSDART00000131309 |
mef2ca
|
myocyte enhancer factor 2ca |
| chr9_-_1978090 | 0.89 |
ENSDART00000082344
|
hoxd11a
|
homeobox D11a |
| chr11_-_22599584 | 0.88 |
ENSDART00000014062
|
myog
|
myogenin |
| chr21_-_8153165 | 0.88 |
ENSDART00000182580
|
CABZ01074363.1
|
|
| chr22_-_15602593 | 0.83 |
ENSDART00000036075
|
tpm4a
|
tropomyosin 4a |
| chr13_+_30912117 | 0.83 |
ENSDART00000133138
|
drgx
|
dorsal root ganglia homeobox |
| chr3_-_28428198 | 0.83 |
ENSDART00000151546
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr24_+_38306010 | 0.82 |
ENSDART00000143184
|
mybpc2b
|
myosin binding protein C, fast type b |
| chr20_-_10120442 | 0.80 |
ENSDART00000144970
|
meis2b
|
Meis homeobox 2b |
| chr6_-_57938043 | 0.80 |
ENSDART00000171073
|
tox2
|
TOX high mobility group box family member 2 |
| chr22_-_15602760 | 0.78 |
ENSDART00000009054
|
tpm4a
|
tropomyosin 4a |
| chr19_-_47452874 | 0.74 |
ENSDART00000025931
|
tfap2e
|
transcription factor AP-2 epsilon |
| chr7_-_31441420 | 0.73 |
ENSDART00000075398
|
cilp
|
cartilage intermediate layer protein, nucleotide pyrophosphohydrolase |
| chr22_-_910926 | 0.71 |
ENSDART00000180075
|
FP016205.1
|
|
| chr19_+_10339538 | 0.70 |
ENSDART00000151808
ENSDART00000151235 |
rcvrn3
|
recoverin 3 |
| chr21_-_36972127 | 0.68 |
ENSDART00000100310
|
dbn1
|
drebrin 1 |
| chr1_-_38195012 | 0.66 |
ENSDART00000020409
|
hand2
|
heart and neural crest derivatives expressed 2 |
| chr19_-_6303099 | 0.66 |
ENSDART00000133371
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr7_+_26224211 | 0.65 |
ENSDART00000173999
|
vgf
|
VGF nerve growth factor inducible |
| chr5_+_57320113 | 0.64 |
ENSDART00000036331
|
atp6v1g1
|
ATPase H+ transporting V1 subunit G1 |
| chr20_+_28434196 | 0.64 |
ENSDART00000034245
|
dpf3
|
D4, zinc and double PHD fingers, family 3 |
| chr22_-_23706771 | 0.63 |
ENSDART00000159771
|
cfhl1
|
complement factor H like 1 |
| chr9_-_33329700 | 0.63 |
ENSDART00000147265
ENSDART00000140039 |
rpl8
|
ribosomal protein L8 |
| chr10_+_21656654 | 0.62 |
ENSDART00000160464
|
pcdh1g2
|
protocadherin 1 gamma 2 |
| chr6_+_12865137 | 0.62 |
ENSDART00000090065
|
fam117ba
|
family with sequence similarity 117, member Ba |
| chr15_-_34567370 | 0.61 |
ENSDART00000099793
|
sostdc1a
|
sclerostin domain containing 1a |
| chr8_+_25351863 | 0.61 |
ENSDART00000034092
|
dnase1l1l
|
deoxyribonuclease I-like 1-like |
| chr3_-_15264698 | 0.59 |
ENSDART00000111948
ENSDART00000142594 |
sez6l2
|
seizure related 6 homolog (mouse)-like 2 |
| chr13_-_9598320 | 0.59 |
ENSDART00000184613
|
cpxm1a
|
carboxypeptidase X (M14 family), member 1a |
| chr3_-_32603191 | 0.58 |
ENSDART00000150997
|
si:ch73-248e21.7
|
si:ch73-248e21.7 |
| chr11_+_21050326 | 0.58 |
ENSDART00000065984
|
zgc:113307
|
zgc:113307 |
| chr7_+_27253063 | 0.58 |
ENSDART00000191138
|
sox6
|
SRY (sex determining region Y)-box 6 |
| chr20_-_49681850 | 0.58 |
ENSDART00000025926
|
col12a1b
|
collagen, type XII, alpha 1b |
| chr11_-_4175773 | 0.58 |
ENSDART00000042221
|
ccdc3b
|
coiled-coil domain containing 3b |
| chr16_-_45209684 | 0.57 |
ENSDART00000184595
|
fxyd1
|
FXYD domain containing ion transport regulator 1 (phospholemman) |
| chr18_+_17583479 | 0.57 |
ENSDART00000186977
ENSDART00000010998 |
slc12a3
|
solute carrier family 12 (sodium/chloride transporter), member 3 |
| chr4_-_22311610 | 0.55 |
ENSDART00000137814
|
hcls1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr1_+_52929185 | 0.54 |
ENSDART00000147683
|
inpp4b
|
inositol polyphosphate-4-phosphatase type II B |
| chr23_+_45282858 | 0.53 |
ENSDART00000162353
|
CABZ01073265.1
|
|
| chr9_-_18568927 | 0.53 |
ENSDART00000084668
|
enox1
|
ecto-NOX disulfide-thiol exchanger 1 |
| chr13_+_30912385 | 0.53 |
ENSDART00000182642
|
drgx
|
dorsal root ganglia homeobox |
| chr4_-_23963838 | 0.52 |
ENSDART00000133433
ENSDART00000132615 ENSDART00000135942 ENSDART00000139439 |
celf2
|
cugbp, Elav-like family member 2 |
| chr7_+_69653981 | 0.51 |
ENSDART00000090165
|
frem1a
|
Fras1 related extracellular matrix 1a |
| chr18_+_8340886 | 0.51 |
ENSDART00000081132
|
cpt1b
|
carnitine palmitoyltransferase 1B (muscle) |
| chr5_-_20205075 | 0.49 |
ENSDART00000051611
|
dao.3
|
D-amino-acid oxidase, tandem duplicate 3 |
| chr9_+_25776971 | 0.49 |
ENSDART00000146011
|
zeb2a
|
zinc finger E-box binding homeobox 2a |
| chr16_+_25535993 | 0.49 |
ENSDART00000077436
|
mylipb
|
myosin regulatory light chain interacting protein b |
| chr16_-_12319822 | 0.48 |
ENSDART00000127453
ENSDART00000184526 |
trpv6
|
transient receptor potential cation channel, subfamily V, member 6 |
| chr9_+_25775816 | 0.48 |
ENSDART00000127834
ENSDART00000189994 |
zeb2a
|
zinc finger E-box binding homeobox 2a |
| chr13_+_29771463 | 0.48 |
ENSDART00000134424
ENSDART00000138332 ENSDART00000134330 ENSDART00000160944 ENSDART00000076992 ENSDART00000160921 |
pax2a
|
paired box 2a |
| chr13_-_1085961 | 0.47 |
ENSDART00000171872
ENSDART00000166598 |
plek
|
pleckstrin |
| chr24_+_792429 | 0.46 |
ENSDART00000082523
|
impa2
|
inositol(myo)-1(or 4)-monophosphatase 2 |
| chr15_-_21014015 | 0.46 |
ENSDART00000144991
|
si:ch211-212c13.10
|
si:ch211-212c13.10 |
| chr18_+_924949 | 0.46 |
ENSDART00000170888
ENSDART00000193163 |
pkma
|
pyruvate kinase M1/2a |
| chr11_+_37201483 | 0.45 |
ENSDART00000160930
ENSDART00000173439 ENSDART00000171273 |
zgc:112265
|
zgc:112265 |
| chr23_+_36095260 | 0.45 |
ENSDART00000127384
|
hoxc9a
|
homeobox C9a |
| chr7_+_20512419 | 0.45 |
ENSDART00000173907
|
si:dkey-19b23.14
|
si:dkey-19b23.14 |
| chr2_+_37227011 | 0.45 |
ENSDART00000126587
ENSDART00000084958 |
samd7
|
sterile alpha motif domain containing 7 |
| chr3_-_19899914 | 0.43 |
ENSDART00000134969
|
rnd2
|
Rho family GTPase 2 |
| chr18_-_898870 | 0.43 |
ENSDART00000151777
ENSDART00000062654 |
parp6a
|
poly (ADP-ribose) polymerase family, member 6a |
| chr21_-_26918901 | 0.43 |
ENSDART00000100685
|
lrfn4a
|
leucine rich repeat and fibronectin type III domain containing 4a |
| chr13_-_30161684 | 0.43 |
ENSDART00000040409
|
ppa1b
|
pyrophosphatase (inorganic) 1b |
| chr11_+_6650966 | 0.43 |
ENSDART00000131236
|
si:dkey-246j7.1
|
si:dkey-246j7.1 |
| chr22_-_11623063 | 0.42 |
ENSDART00000145653
|
gcga
|
glucagon a |
| chr2_-_34555945 | 0.42 |
ENSDART00000056671
|
brinp2
|
bone morphogenetic protein/retinoic acid inducible neural-specific 2 |
| chr14_-_33105434 | 0.41 |
ENSDART00000163795
|
dlg3
|
discs, large homolog 3 (Drosophila) |
| chr18_+_31410652 | 0.41 |
ENSDART00000098504
|
def8
|
differentially expressed in FDCP 8 homolog (mouse) |
| chr15_+_32268790 | 0.41 |
ENSDART00000154457
|
fhdc4
|
FH2 domain containing 4 |
| chr9_-_23383131 | 0.40 |
ENSDART00000101685
|
sh3bp4
|
SH3-domain binding protein 4 |
| chr10_-_7974155 | 0.40 |
ENSDART00000147368
ENSDART00000075524 |
osbp2
|
oxysterol binding protein 2 |
| chr8_+_6967108 | 0.39 |
ENSDART00000004588
|
asic1a
|
acid-sensing (proton-gated) ion channel 1a |
| chr13_+_23162447 | 0.39 |
ENSDART00000180209
|
sorbs1
|
sorbin and SH3 domain containing 1 |
| chr4_-_5595237 | 0.39 |
ENSDART00000109854
|
vegfab
|
vascular endothelial growth factor Ab |
| chr5_-_22882210 | 0.38 |
ENSDART00000182232
|
si:ch211-26b3.4
|
si:ch211-26b3.4 |
| chr4_+_11479705 | 0.38 |
ENSDART00000019458
ENSDART00000150587 ENSDART00000139370 ENSDART00000135826 |
asb13a.1
|
ankyrin repeat and SOCS box containing 13a, tandem duplicate 1 |
| chr11_-_10828539 | 0.37 |
ENSDART00000040180
|
tbr1a
|
T-box, brain, 1a |
| chr14_+_19258702 | 0.37 |
ENSDART00000187087
ENSDART00000005738 |
slitrk2
|
SLIT and NTRK-like family, member 2 |
| chr20_+_34151670 | 0.37 |
ENSDART00000152870
ENSDART00000010329 ENSDART00000145852 |
arpc5b
|
actin related protein 2/3 complex, subunit 5B |
| chr6_-_32093830 | 0.37 |
ENSDART00000017695
|
foxd3
|
forkhead box D3 |
| chr22_-_23000815 | 0.36 |
ENSDART00000137111
|
ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr1_+_14073891 | 0.35 |
ENSDART00000021693
|
ank2a
|
ankyrin 2a, neuronal |
| chr20_+_23173710 | 0.35 |
ENSDART00000074172
|
sgcb
|
sarcoglycan, beta (dystrophin-associated glycoprotein) |
| chr13_-_33667922 | 0.35 |
ENSDART00000141631
|
si:dkey-76k16.5
|
si:dkey-76k16.5 |
| chr8_-_53198154 | 0.34 |
ENSDART00000083416
|
gabrd
|
gamma-aminobutyric acid (GABA) A receptor, delta |
| chr20_-_38610360 | 0.33 |
ENSDART00000168783
ENSDART00000164638 |
slc30a2
|
solute carrier family 30 (zinc transporter), member 2 |
| chr4_+_14727018 | 0.32 |
ENSDART00000124189
|
cmasa
|
cytidine monophosphate N-acetylneuraminic acid synthetase a |
| chr22_-_29586608 | 0.32 |
ENSDART00000059869
|
adra2a
|
adrenoceptor alpha 2A |
| chr20_-_22476255 | 0.32 |
ENSDART00000103510
|
pdgfra
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr20_+_25225112 | 0.32 |
ENSDART00000153088
ENSDART00000127291 ENSDART00000130494 |
moxd1
|
monooxygenase, DBH-like 1 |
| chr4_+_612363 | 0.31 |
ENSDART00000049154
|
pthlha
|
parathyroid hormone-like hormone a |
| chr18_+_7286788 | 0.30 |
ENSDART00000022998
|
ANO2 (1 of many)
|
si:ch73-86n2.1 |
| chr9_-_16853462 | 0.29 |
ENSDART00000160273
|
CT573248.2
|
|
| chr5_+_36654817 | 0.29 |
ENSDART00000131339
|
capns1a
|
calpain, small subunit 1 a |
| chr18_+_30028637 | 0.28 |
ENSDART00000139750
|
si:ch211-220f16.1
|
si:ch211-220f16.1 |
| chr18_+_26899316 | 0.28 |
ENSDART00000050230
|
tspan3a
|
tetraspanin 3a |
| chr2_-_45027754 | 0.27 |
ENSDART00000168469
|
vwa5b2
|
von Willebrand factor A domain containing 5B2 |
| chr8_+_25352268 | 0.27 |
ENSDART00000187829
|
dnase1l1l
|
deoxyribonuclease I-like 1-like |
| chr8_-_19216180 | 0.27 |
ENSDART00000146162
|
si:ch73-222f22.2
|
si:ch73-222f22.2 |
| chr4_-_21652812 | 0.26 |
ENSDART00000174400
|
rps16
|
ribosomal protein S16 |
| chr13_+_17702522 | 0.26 |
ENSDART00000057913
|
si:dkey-27m7.4
|
si:dkey-27m7.4 |
| chr6_+_20647155 | 0.26 |
ENSDART00000193477
|
slc19a1
|
solute carrier family 19 (folate transporter), member 1 |
| chr4_+_14727212 | 0.26 |
ENSDART00000158094
|
cmasa
|
cytidine monophosphate N-acetylneuraminic acid synthetase a |
| chr1_-_58036509 | 0.25 |
ENSDART00000081122
|
COLGALT1
|
si:ch211-114l13.7 |
| chr20_-_39273987 | 0.25 |
ENSDART00000127173
|
clu
|
clusterin |
| chr5_+_33339762 | 0.25 |
ENSDART00000026085
|
ptges
|
prostaglandin E synthase |
| chr1_-_59077650 | 0.24 |
ENSDART00000043516
|
MFAP4 (1 of many)
|
si:zfos-2330d3.1 |
| chr16_+_45739193 | 0.24 |
ENSDART00000184852
ENSDART00000156851 ENSDART00000154704 |
paqr6
|
progestin and adipoQ receptor family member VI |
| chr6_-_957830 | 0.24 |
ENSDART00000090019
ENSDART00000184286 |
zeb2b
|
zinc finger E-box binding homeobox 2b |
| chr24_+_119680 | 0.24 |
ENSDART00000061973
|
tgfbr1b
|
transforming growth factor, beta receptor 1 b |
| chr2_-_34483597 | 0.24 |
ENSDART00000133224
|
brinp2
|
bone morphogenetic protein/retinoic acid inducible neural-specific 2 |
| chr3_-_32169754 | 0.24 |
ENSDART00000179010
|
tnnt1
|
troponin T type 1 (skeletal, slow) |
| chr20_+_46183505 | 0.23 |
ENSDART00000060799
|
taar13b
|
trace amine associated receptor 13b |
| chr17_-_22552678 | 0.23 |
ENSDART00000079401
|
si:ch211-125o16.4
|
si:ch211-125o16.4 |
| chr6_+_11249706 | 0.23 |
ENSDART00000186547
ENSDART00000193287 |
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr10_-_42131408 | 0.23 |
ENSDART00000076693
|
stambpa
|
STAM binding protein a |
| chr20_-_39273505 | 0.22 |
ENSDART00000153114
|
clu
|
clusterin |
| chr5_-_69548986 | 0.22 |
ENSDART00000097250
ENSDART00000163390 ENSDART00000166117 ENSDART00000167625 ENSDART00000191777 |
kiss1rb
|
KISS1 receptor b |
| chr14_+_46274611 | 0.22 |
ENSDART00000134363
|
cabp2b
|
calcium binding protein 2b |
| chr3_+_25166805 | 0.22 |
ENSDART00000077493
|
TST
|
zgc:162544 |
| chr14_+_17197132 | 0.22 |
ENSDART00000054598
|
rtn4rl2b
|
reticulon 4 receptor-like 2b |
| chr19_+_19511394 | 0.22 |
ENSDART00000172410
|
jazf1a
|
JAZF zinc finger 1a |
| chr10_+_14210667 | 0.22 |
ENSDART00000138854
|
si:dkey-286h21.1
|
si:dkey-286h21.1 |
| chr5_-_65000312 | 0.21 |
ENSDART00000192893
|
ANXA1 (1 of many)
|
zgc:110283 |
| chr1_-_23598657 | 0.21 |
ENSDART00000144348
ENSDART00000142641 ENSDART00000143588 |
si:dkeyp-26a9.7
|
si:dkeyp-26a9.7 |
| chr12_+_9551667 | 0.21 |
ENSDART00000048493
ENSDART00000166178 |
p4ha1a
|
prolyl 4-hydroxylase, alpha polypeptide I a |
| chr4_+_31405646 | 0.21 |
ENSDART00000128732
|
si:rp71-5o12.3
|
si:rp71-5o12.3 |
| chr2_-_32551178 | 0.20 |
ENSDART00000145603
|
smarcd3a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3a |
| chr24_-_21819010 | 0.20 |
ENSDART00000091096
|
CR352265.1
|
|
| chr23_-_44916800 | 0.20 |
ENSDART00000150076
|
fhdc5
|
FH2 domain containing 5 |
| chr5_-_54197084 | 0.20 |
ENSDART00000163640
|
grk1b
|
G protein-coupled receptor kinase 1 b |
| chr18_-_48558420 | 0.20 |
ENSDART00000058987
|
kcnj1a.1
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 1 |
| chr15_+_36966369 | 0.20 |
ENSDART00000163622
|
kirrel3l
|
kirre like nephrin family adhesion molecule 3, like |
| chr8_-_2289264 | 0.20 |
ENSDART00000189397
|
plat
|
plasminogen activator, tissue |
| chr22_+_38164486 | 0.19 |
ENSDART00000137521
|
tm4sf18
|
transmembrane 4 L six family member 18 |
| chr24_+_20920563 | 0.19 |
ENSDART00000037224
|
cst14a.2
|
cystatin 14a, tandem duplicate 2 |
| chr11_+_30981777 | 0.19 |
ENSDART00000148949
|
cacna1ab
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit, b |
| chr17_-_2386569 | 0.19 |
ENSDART00000121614
|
PLCB2
|
phospholipase C beta 2 |
| chr7_+_72469724 | 0.19 |
ENSDART00000187629
|
HECTD4
|
HECT domain E3 ubiquitin protein ligase 4 |
| chr20_-_25436389 | 0.18 |
ENSDART00000153266
|
itsn2a
|
intersectin 2a |
| chr9_-_21918963 | 0.18 |
ENSDART00000090782
|
lmo7a
|
LIM domain 7a |
| chr9_-_41277347 | 0.18 |
ENSDART00000181213
|
glsb
|
glutaminase b |
| chr23_-_21797517 | 0.18 |
ENSDART00000110041
|
lrrc38a
|
leucine rich repeat containing 38a |
| chr2_-_14387335 | 0.18 |
ENSDART00000189332
ENSDART00000164786 ENSDART00000188572 |
sgip1b
|
SH3-domain GRB2-like (endophilin) interacting protein 1b |
| chr5_-_52921532 | 0.17 |
ENSDART00000160566
|
CU695117.1
|
|
| chr2_+_45548890 | 0.17 |
ENSDART00000113994
|
fndc7a
|
fibronectin type III domain containing 7a |
| chr7_-_5316901 | 0.16 |
ENSDART00000181505
ENSDART00000124367 |
si:cabz01074946.1
|
si:cabz01074946.1 |
| chr6_+_11250033 | 0.16 |
ENSDART00000065411
ENSDART00000132677 |
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr15_-_4094888 | 0.16 |
ENSDART00000166307
|
TM4SF19
|
si:dkey-83h2.3 |
| chr1_-_44638058 | 0.16 |
ENSDART00000081835
|
slc43a1b
|
solute carrier family 43 (amino acid system L transporter), member 1b |
| chr7_+_2228276 | 0.16 |
ENSDART00000064294
|
si:dkey-187j14.4
|
si:dkey-187j14.4 |
| chr7_+_48870496 | 0.16 |
ENSDART00000009642
|
igf2a
|
insulin-like growth factor 2a |
| chr1_-_28831848 | 0.15 |
ENSDART00000148536
|
GK3P
|
zgc:172295 |
| chr17_-_8173380 | 0.15 |
ENSDART00000148520
|
syne1b
|
spectrin repeat containing, nuclear envelope 1b |
| chr15_+_16343073 | 0.15 |
ENSDART00000029063
|
p2rx8
|
purinergic receptor P2X, ligand-gated ion channel, 8 |
| chr3_-_34180364 | 0.15 |
ENSDART00000151819
ENSDART00000003133 |
yipf2
|
Yip1 domain family, member 2 |
| chr24_+_9003998 | 0.15 |
ENSDART00000179656
ENSDART00000191314 |
CR318624.2
|
|
| chr20_+_18551657 | 0.15 |
ENSDART00000147001
|
si:dkeyp-72h1.1
|
si:dkeyp-72h1.1 |
| chr3_-_55197881 | 0.14 |
ENSDART00000179903
|
kank2
|
KN motif and ankyrin repeat domains 2 |
| chr1_-_40086978 | 0.14 |
ENSDART00000136990
|
si:dkey-117m1.4
|
si:dkey-117m1.4 |
| chr23_-_43393589 | 0.14 |
ENSDART00000183592
|
zgc:174862
|
zgc:174862 |
| chr5_+_51026563 | 0.14 |
ENSDART00000050988
|
gcnt4a
|
glucosaminyl (N-acetyl) transferase 4, core 2, a |
| chr3_+_34180835 | 0.13 |
ENSDART00000055252
|
timm29
|
translocase of inner mitochondrial membrane 29 |
| chr9_+_25776194 | 0.13 |
ENSDART00000144499
|
zeb2a
|
zinc finger E-box binding homeobox 2a |
| chr19_+_816208 | 0.13 |
ENSDART00000093304
|
nrm
|
nurim |
| chr24_+_26887487 | 0.13 |
ENSDART00000189425
|
CR376848.1
|
|
| chr25_+_28893615 | 0.13 |
ENSDART00000156994
ENSDART00000075151 |
amn1
|
antagonist of mitotic exit network 1 homolog (S. cerevisiae) |
| chr15_-_31364729 | 0.12 |
ENSDART00000185386
|
or111-2
|
odorant receptor, family D, subfamily 111, member 2 |
| chr4_-_287425 | 0.12 |
ENSDART00000159128
|
echdc3
|
enoyl CoA hydratase domain containing 3 |
| chr3_+_30968176 | 0.12 |
ENSDART00000186266
|
prf1.9
|
perforin 1.9 |
| chr13_+_32148338 | 0.12 |
ENSDART00000188591
|
osr1
|
odd-skipped related transciption factor 1 |
| chr14_-_8625845 | 0.11 |
ENSDART00000054760
|
zgc:162144
|
zgc:162144 |
| chr3_-_17716322 | 0.11 |
ENSDART00000192664
|
CABZ01018956.1
|
|
| chr7_+_26326462 | 0.11 |
ENSDART00000173515
|
zanl
|
zonadhesin, like |
| chr25_-_28768489 | 0.11 |
ENSDART00000088315
|
washc4
|
WASH complex subunit 4 |
| chr22_+_10010292 | 0.10 |
ENSDART00000180096
|
BX324216.4
|
|
| chr24_-_32025637 | 0.10 |
ENSDART00000180448
ENSDART00000159034 |
rsu1
|
Ras suppressor protein 1 |
| chr9_+_49712868 | 0.10 |
ENSDART00000192969
ENSDART00000183310 |
CSRNP3
|
cysteine and serine rich nuclear protein 3 |
| chr2_+_20332044 | 0.10 |
ENSDART00000112131
|
plppr4a
|
phospholipid phosphatase related 4a |
Network of associatons between targets according to the STRING database.
First level regulatory network of lhx8a+lhx8b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.9 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.3 | 0.9 | GO:0036076 | ligamentous ossification(GO:0036076) |
| 0.2 | 0.9 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.2 | 1.1 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.1 | 0.7 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.1 | 0.7 | GO:0071908 | determination of intestine left/right asymmetry(GO:0071908) |
| 0.1 | 1.7 | GO:0006603 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.1 | 0.5 | GO:0046436 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.1 | 0.5 | GO:0045023 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.1 | 0.9 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.1 | 0.5 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.1 | 0.3 | GO:0046333 | octopamine biosynthetic process(GO:0006589) dopamine catabolic process(GO:0042420) norepinephrine biosynthetic process(GO:0042421) octopamine metabolic process(GO:0046333) |
| 0.1 | 1.0 | GO:0034434 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.1 | 0.5 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.1 | 0.3 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.1 | 0.3 | GO:0098838 | folic acid transport(GO:0015884) drug transport(GO:0015893) methotrexate transport(GO:0051958) reduced folate transmembrane transport(GO:0098838) |
| 0.1 | 0.3 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 | 0.9 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.1 | 1.9 | GO:0006942 | regulation of striated muscle contraction(GO:0006942) |
| 0.1 | 0.4 | GO:0097066 | response to thyroid hormone(GO:0097066) |
| 0.1 | 1.0 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 0.7 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 0.3 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.1 | 0.3 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.1 | 0.4 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
| 0.1 | 0.8 | GO:0003209 | cardiac atrium morphogenesis(GO:0003209) |
| 0.1 | 0.5 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.1 | 0.6 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.1 | 0.1 | GO:0048389 | intermediate mesoderm development(GO:0048389) |
| 0.1 | 0.6 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.1 | 1.0 | GO:0001990 | regulation of systemic arterial blood pressure by hormone(GO:0001990) |
| 0.1 | 0.4 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.1 | 1.6 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 0.6 | GO:0039023 | pronephric duct morphogenesis(GO:0039023) |
| 0.1 | 0.9 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.0 | 0.2 | GO:0014909 | smooth muscle cell migration(GO:0014909) |
| 0.0 | 0.2 | GO:0003010 | voluntary skeletal muscle contraction(GO:0003010) twitch skeletal muscle contraction(GO:0014721) slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 3.7 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 1.0 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.0 | 0.1 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.5 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.0 | 0.6 | GO:0048679 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.0 | 0.6 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.0 | 0.3 | GO:0097531 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.5 | GO:0071545 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) inositol phosphate catabolic process(GO:0071545) |
| 0.0 | 0.3 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.2 | GO:0032986 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.2 | GO:0006537 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.0 | 1.0 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.1 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.0 | 0.8 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.0 | 0.2 | GO:0039022 | pronephric duct development(GO:0039022) |
| 0.0 | 0.5 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.9 | GO:0006956 | complement activation(GO:0006956) |
| 0.0 | 0.1 | GO:0045852 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.0 | 0.2 | GO:0046626 | regulation of insulin receptor signaling pathway(GO:0046626) |
| 0.0 | 0.2 | GO:0033198 | response to ATP(GO:0033198) |
| 0.0 | 0.1 | GO:0019483 | beta-alanine metabolic process(GO:0019482) beta-alanine biosynthetic process(GO:0019483) |
| 0.0 | 0.2 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.0 | 1.3 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 0.4 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.1 | GO:0001516 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 | 0.6 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.2 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 1.0 | GO:0016079 | synaptic vesicle exocytosis(GO:0016079) |
| 0.0 | 0.4 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 | 1.3 | GO:0021782 | glial cell development(GO:0021782) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.2 | 1.9 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 0.9 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 2.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 1.2 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.5 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.2 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.8 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.6 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.4 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 2.8 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.3 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.3 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.1 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.0 | 0.7 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.6 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.9 | GO:0019863 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.2 | 1.0 | GO:0004772 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.2 | 1.0 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.2 | 1.9 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.2 | 0.5 | GO:0052834 | inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.1 | 1.7 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.1 | 0.5 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.1 | 0.9 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.3 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.1 | 0.4 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.1 | 0.3 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.1 | 1.2 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 0.4 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 0.9 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.1 | 0.3 | GO:0008518 | reduced folate carrier activity(GO:0008518) methotrexate transporter activity(GO:0015350) |
| 0.1 | 0.4 | GO:0031769 | glucagon receptor binding(GO:0031769) |
| 0.1 | 0.3 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.4 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 0.6 | GO:0036122 | BMP binding(GO:0036122) |
| 0.1 | 0.5 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.1 | 0.5 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.5 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 0.6 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.1 | 0.3 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.1 | 2.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.6 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 3.9 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 1.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0050254 | rhodopsin kinase activity(GO:0050254) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.3 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.2 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.0 | 0.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.9 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.5 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.4 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.2 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 2.5 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.0 | 0.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.2 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.6 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 4.2 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.8 | GO:0031490 | chromatin DNA binding(GO:0031490) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.4 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.5 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.4 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.4 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 2.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.3 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.5 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.9 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 0.4 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.5 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.3 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.9 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.0 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |