Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
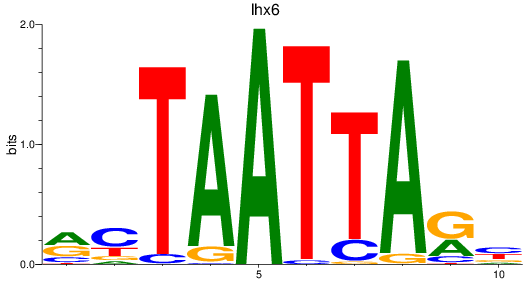
Results for lhx6
Z-value: 0.83
Transcription factors associated with lhx6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
lhx6
|
ENSDARG00000006896 | LIM homeobox 6 |
|
lhx6
|
ENSDARG00000112520 | LIM homeobox 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| lhx6 | dr11_v1_chr10_+_9372702_9372702 | 0.81 | 7.5e-03 | Click! |
Activity profile of lhx6 motif
Sorted Z-values of lhx6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_15433518 | 2.22 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr17_+_15433671 | 2.16 |
ENSDART00000149568
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr3_-_61162750 | 1.52 |
ENSDART00000055064
|
pvalb8
|
parvalbumin 8 |
| chr9_-_22099536 | 1.48 |
ENSDART00000101923
|
CR391987.1
|
|
| chr4_-_9891874 | 1.32 |
ENSDART00000067193
|
adm2a
|
adrenomedullin 2a |
| chr20_-_9436521 | 1.29 |
ENSDART00000133000
|
zgc:101840
|
zgc:101840 |
| chr5_-_64831207 | 1.17 |
ENSDART00000144816
|
lix1
|
limb and CNS expressed 1 |
| chr3_-_46811611 | 1.12 |
ENSDART00000134092
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr20_+_26880668 | 1.11 |
ENSDART00000077769
|
serpinb1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
| chr12_-_35830625 | 1.07 |
ENSDART00000180028
|
CU459056.1
|
|
| chr8_+_16025554 | 1.05 |
ENSDART00000110171
|
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr20_-_48485354 | 1.04 |
ENSDART00000124040
ENSDART00000148437 |
insm1a
|
insulinoma-associated 1a |
| chr22_-_10121880 | 1.03 |
ENSDART00000002348
|
rdh5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr24_-_21923930 | 1.02 |
ENSDART00000131944
|
tagln3b
|
transgelin 3b |
| chr15_-_23376541 | 0.91 |
ENSDART00000078570
|
c1qtnf5
|
C1q and TNF related 5 |
| chr15_+_9072821 | 0.88 |
ENSDART00000154463
|
si:dkey-202g17.3
|
si:dkey-202g17.3 |
| chr5_-_41307550 | 0.88 |
ENSDART00000143446
|
npr3
|
natriuretic peptide receptor 3 |
| chr5_+_64732270 | 0.88 |
ENSDART00000134241
|
olfm1a
|
olfactomedin 1a |
| chr5_+_64732036 | 0.87 |
ENSDART00000073950
|
olfm1a
|
olfactomedin 1a |
| chr16_+_31804590 | 0.87 |
ENSDART00000167321
|
wnt4b
|
wingless-type MMTV integration site family, member 4b |
| chr1_-_19845378 | 0.81 |
ENSDART00000139314
ENSDART00000132958 ENSDART00000147502 |
grhprb
|
glyoxylate reductase/hydroxypyruvate reductase b |
| chr11_+_38280454 | 0.75 |
ENSDART00000171496
|
CDK18
|
si:dkey-166c18.1 |
| chr13_+_255067 | 0.73 |
ENSDART00000102505
|
foxg1d
|
forkhead box G1d |
| chr13_+_19322686 | 0.73 |
ENSDART00000058036
|
emx2
|
empty spiracles homeobox 2 |
| chr15_+_47903864 | 0.72 |
ENSDART00000063835
|
otx5
|
orthodenticle homolog 5 |
| chr21_-_22827548 | 0.71 |
ENSDART00000079161
|
angptl5
|
angiopoietin-like 5 |
| chr4_+_21129752 | 0.71 |
ENSDART00000169764
|
syt1a
|
synaptotagmin Ia |
| chr5_-_51619742 | 0.70 |
ENSDART00000188537
|
otpb
|
orthopedia homeobox b |
| chr11_-_39044595 | 0.69 |
ENSDART00000065461
|
cldn19
|
claudin 19 |
| chr22_-_15593824 | 0.68 |
ENSDART00000123125
|
tpm4a
|
tropomyosin 4a |
| chr16_+_46111849 | 0.66 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr7_+_30787903 | 0.65 |
ENSDART00000174000
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr21_+_28958471 | 0.65 |
ENSDART00000144331
ENSDART00000005929 |
ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr3_-_46817499 | 0.65 |
ENSDART00000013717
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr13_-_31622195 | 0.64 |
ENSDART00000057432
|
six1a
|
SIX homeobox 1a |
| chr7_-_35708450 | 0.63 |
ENSDART00000193886
|
irx5a
|
iroquois homeobox 5a |
| chr1_-_50859053 | 0.62 |
ENSDART00000132779
ENSDART00000137648 |
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr9_+_25776971 | 0.61 |
ENSDART00000146011
|
zeb2a
|
zinc finger E-box binding homeobox 2a |
| chr3_-_53533128 | 0.60 |
ENSDART00000183591
|
notch3
|
notch 3 |
| chr14_+_14662116 | 0.60 |
ENSDART00000161693
|
cetn2
|
centrin, EF-hand protein, 2 |
| chr12_+_20352400 | 0.60 |
ENSDART00000066383
|
hbae5
|
hemoglobin, alpha embryonic 5 |
| chr5_+_56268436 | 0.60 |
ENSDART00000021159
|
lhx1b
|
LIM homeobox 1b |
| chr1_+_25801648 | 0.59 |
ENSDART00000129471
|
gucy1b1
|
guanylate cyclase 1 soluble subunit beta 1 |
| chr13_+_35339182 | 0.59 |
ENSDART00000019323
|
jag1b
|
jagged 1b |
| chr13_-_31435137 | 0.59 |
ENSDART00000057441
|
rtn1a
|
reticulon 1a |
| chr18_+_30847237 | 0.59 |
ENSDART00000012374
|
foxf1
|
forkhead box F1 |
| chr9_+_34641237 | 0.58 |
ENSDART00000133996
|
shox
|
short stature homeobox |
| chr22_-_13350240 | 0.58 |
ENSDART00000154095
ENSDART00000155118 |
si:ch211-227m13.1
|
si:ch211-227m13.1 |
| chr18_+_9171778 | 0.58 |
ENSDART00000101192
|
sema3d
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D |
| chr9_+_30108641 | 0.57 |
ENSDART00000060174
|
jagn1a
|
jagunal homolog 1a |
| chr3_-_50443607 | 0.57 |
ENSDART00000074036
|
rcvrna
|
recoverin a |
| chr3_-_43356082 | 0.56 |
ENSDART00000171213
|
uncx
|
UNC homeobox |
| chr12_-_4781801 | 0.56 |
ENSDART00000167490
ENSDART00000121718 |
mapta
|
microtubule-associated protein tau a |
| chr3_-_12930217 | 0.55 |
ENSDART00000166322
|
pdgfab
|
platelet-derived growth factor alpha polypeptide b |
| chr20_-_16171297 | 0.55 |
ENSDART00000012476
|
coa7
|
cytochrome c oxidase assembly factor 7 |
| chr7_-_28148310 | 0.54 |
ENSDART00000044208
|
lmo1
|
LIM domain only 1 |
| chr13_-_24311628 | 0.54 |
ENSDART00000004420
|
rab4a
|
RAB4a, member RAS oncogene family |
| chr2_-_32558795 | 0.54 |
ENSDART00000140026
|
smarcd3a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3a |
| chr3_+_32553714 | 0.53 |
ENSDART00000165638
|
pax10
|
paired box 10 |
| chr18_+_16744307 | 0.51 |
ENSDART00000179872
ENSDART00000133490 |
lyve1b
|
lymphatic vessel endothelial hyaluronic receptor 1b |
| chr2_+_35728033 | 0.51 |
ENSDART00000002094
|
ankrd45
|
ankyrin repeat domain 45 |
| chr2_+_50608099 | 0.51 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr11_-_42554290 | 0.51 |
ENSDART00000130573
|
atp6ap1la
|
ATPase H+ transporting accessory protein 1 like a |
| chr5_-_63515210 | 0.50 |
ENSDART00000022348
|
prdm12b
|
PR domain containing 12b |
| chr8_+_29986265 | 0.50 |
ENSDART00000148258
|
ptch1
|
patched 1 |
| chr4_-_4834347 | 0.48 |
ENSDART00000141803
|
coa6
|
cytochrome c oxidase assembly factor 6 |
| chr4_-_17629444 | 0.48 |
ENSDART00000108814
|
nrip2
|
nuclear receptor interacting protein 2 |
| chr16_-_42894628 | 0.47 |
ENSDART00000045600
|
hfe2
|
hemochromatosis type 2 |
| chr13_-_29420885 | 0.47 |
ENSDART00000024225
|
chata
|
choline O-acetyltransferase a |
| chr4_-_4834617 | 0.45 |
ENSDART00000141539
|
coa6
|
cytochrome c oxidase assembly factor 6 |
| chr4_+_11384891 | 0.45 |
ENSDART00000092381
ENSDART00000186577 ENSDART00000191054 ENSDART00000191584 |
pcloa
|
piccolo presynaptic cytomatrix protein a |
| chr17_-_36896560 | 0.44 |
ENSDART00000045287
|
mapre3a
|
microtubule-associated protein, RP/EB family, member 3a |
| chr3_+_45687266 | 0.44 |
ENSDART00000131652
|
gpr146
|
G protein-coupled receptor 146 |
| chr3_-_60175470 | 0.43 |
ENSDART00000156597
|
si:ch73-364h19.1
|
si:ch73-364h19.1 |
| chr14_-_4145594 | 0.43 |
ENSDART00000077348
|
casp3b
|
caspase 3, apoptosis-related cysteine peptidase b |
| chr20_+_37294112 | 0.43 |
ENSDART00000076293
ENSDART00000140450 |
cx23
|
connexin 23 |
| chr15_-_22074315 | 0.42 |
ENSDART00000149830
|
drd2a
|
dopamine receptor D2a |
| chr19_+_233143 | 0.41 |
ENSDART00000175273
|
syngap1a
|
synaptic Ras GTPase activating protein 1a |
| chr1_-_46981134 | 0.40 |
ENSDART00000130607
|
pknox1.2
|
pbx/knotted 1 homeobox 1.2 |
| chr17_-_13007484 | 0.39 |
ENSDART00000156812
|
si:dkeyp-33b5.4
|
si:dkeyp-33b5.4 |
| chr23_+_28582865 | 0.39 |
ENSDART00000020296
|
l1cama
|
L1 cell adhesion molecule, paralog a |
| chr10_+_21726853 | 0.39 |
ENSDART00000162869
|
pcdh1g14
|
protocadherin 1 gamma 14 |
| chr19_+_19786117 | 0.39 |
ENSDART00000167757
ENSDART00000163546 |
hoxa1a
|
homeobox A1a |
| chr8_-_33381913 | 0.39 |
ENSDART00000076420
|
lmx1bb
|
LIM homeobox transcription factor 1, beta b |
| chr1_-_45049603 | 0.39 |
ENSDART00000023336
|
rps6
|
ribosomal protein S6 |
| chr20_-_46362606 | 0.39 |
ENSDART00000153087
|
bmf2
|
BCL2 modifying factor 2 |
| chr24_+_16547035 | 0.39 |
ENSDART00000164319
|
sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr14_+_17376940 | 0.38 |
ENSDART00000054590
ENSDART00000010148 |
spon2b
|
spondin 2b, extracellular matrix protein |
| chr7_-_49594995 | 0.38 |
ENSDART00000174161
ENSDART00000109147 |
brsk2b
|
BR serine/threonine kinase 2b |
| chr8_-_15129573 | 0.37 |
ENSDART00000142358
|
bcar3
|
BCAR3, NSP family adaptor protein |
| chr16_+_21330634 | 0.37 |
ENSDART00000191285
ENSDART00000183267 |
osbpl3b
|
oxysterol binding protein-like 3b |
| chr1_-_16665044 | 0.37 |
ENSDART00000040434
|
asah1b
|
N-acylsphingosine amidohydrolase (acid ceramidase) 1b |
| chr15_+_23799461 | 0.37 |
ENSDART00000154885
|
si:ch211-167j9.4
|
si:ch211-167j9.4 |
| chr6_-_39270851 | 0.37 |
ENSDART00000148839
|
arhgef25b
|
Rho guanine nucleotide exchange factor (GEF) 25b |
| chr16_-_26296477 | 0.37 |
ENSDART00000157553
|
erfl1
|
Ets2 repressor factor like 1 |
| chr1_-_31534089 | 0.35 |
ENSDART00000007770
|
lbx1b
|
ladybird homeobox 1b |
| chr21_+_30937690 | 0.35 |
ENSDART00000022562
|
rhogb
|
ras homolog family member Gb |
| chr2_-_30668580 | 0.35 |
ENSDART00000087270
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr6_+_39232245 | 0.35 |
ENSDART00000187351
|
b4galnt1b
|
beta-1,4-N-acetyl-galactosaminyl transferase 1b |
| chr14_-_33105434 | 0.35 |
ENSDART00000163795
|
dlg3
|
discs, large homolog 3 (Drosophila) |
| chr23_+_11285662 | 0.34 |
ENSDART00000111028
|
chl1a
|
cell adhesion molecule L1-like a |
| chr18_-_20869175 | 0.34 |
ENSDART00000090079
|
synm
|
synemin, intermediate filament protein |
| chr23_+_31107685 | 0.34 |
ENSDART00000103448
|
tbx18
|
T-box 18 |
| chr3_+_56645710 | 0.33 |
ENSDART00000193978
|
CR759836.1
|
|
| chr7_-_31759602 | 0.33 |
ENSDART00000113467
|
igdcc4
|
immunoglobulin superfamily, DCC subclass, member 4 |
| chr24_+_6107901 | 0.33 |
ENSDART00000156419
|
si:ch211-37e10.2
|
si:ch211-37e10.2 |
| chr5_-_69004007 | 0.33 |
ENSDART00000137443
|
ank1a
|
ankyrin 1, erythrocytic a |
| chr23_+_40460333 | 0.32 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr20_+_539852 | 0.32 |
ENSDART00000185994
|
dse
|
dermatan sulfate epimerase |
| chr13_+_27232848 | 0.32 |
ENSDART00000138043
|
rin2
|
Ras and Rab interactor 2 |
| chr5_-_48268049 | 0.32 |
ENSDART00000187454
|
mef2cb
|
myocyte enhancer factor 2cb |
| chr9_+_38372216 | 0.32 |
ENSDART00000141895
|
plcd4b
|
phospholipase C, delta 4b |
| chr19_+_22062202 | 0.31 |
ENSDART00000100181
|
sall3b
|
spalt-like transcription factor 3b |
| chr9_-_19161982 | 0.31 |
ENSDART00000081878
|
pou1f1
|
POU class 1 homeobox 1 |
| chr3_-_56541723 | 0.31 |
ENSDART00000156398
ENSDART00000050576 ENSDART00000184874 |
si:ch211-189a21.1
cyth1a
|
si:ch211-189a21.1 cytohesin 1a |
| chr18_-_48547564 | 0.31 |
ENSDART00000138607
|
kcnj1a.1
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 1 |
| chr3_-_23406964 | 0.31 |
ENSDART00000114723
|
rapgefl1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr14_-_858985 | 0.30 |
ENSDART00000148687
ENSDART00000149375 |
slc34a1a
|
solute carrier family 34 (type II sodium/phosphate cotransporter), member 1a |
| chr8_+_25902170 | 0.30 |
ENSDART00000193130
|
rhoab
|
ras homolog gene family, member Ab |
| chr24_+_17269849 | 0.30 |
ENSDART00000017605
|
spag6
|
sperm associated antigen 6 |
| chr15_+_1796313 | 0.30 |
ENSDART00000126253
|
fam124b
|
family with sequence similarity 124B |
| chr22_+_5176255 | 0.30 |
ENSDART00000092647
|
cers1
|
ceramide synthase 1 |
| chr20_-_22476255 | 0.30 |
ENSDART00000103510
|
pdgfra
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr6_-_6487876 | 0.29 |
ENSDART00000137642
|
cep170ab
|
centrosomal protein 170Ab |
| chr8_-_25336589 | 0.29 |
ENSDART00000009682
|
atp5pb
|
ATP synthase peripheral stalk-membrane subunit b |
| chr18_+_27738349 | 0.29 |
ENSDART00000187816
|
tspan18b
|
tetraspanin 18b |
| chr24_+_19591893 | 0.29 |
ENSDART00000152026
|
slco5a1a
|
solute carrier organic anion transporter family member 5A1a |
| chr7_+_27834130 | 0.29 |
ENSDART00000052656
|
rras2
|
RAS related 2 |
| chr12_+_20587179 | 0.28 |
ENSDART00000170127
|
arsg
|
arylsulfatase G |
| chr4_+_12612145 | 0.28 |
ENSDART00000181201
|
lmo3
|
LIM domain only 3 |
| chr25_-_6049339 | 0.28 |
ENSDART00000075184
|
snx1a
|
sorting nexin 1a |
| chr7_-_30174882 | 0.27 |
ENSDART00000110409
|
frmd5
|
FERM domain containing 5 |
| chr22_+_5176693 | 0.27 |
ENSDART00000160927
|
cers1
|
ceramide synthase 1 |
| chr10_+_36026576 | 0.27 |
ENSDART00000193786
|
hmgb1a
|
high mobility group box 1a |
| chr10_-_27049170 | 0.27 |
ENSDART00000143451
|
cnih2
|
cornichon family AMPA receptor auxiliary protein 2 |
| chr10_-_11385155 | 0.26 |
ENSDART00000064214
|
plac8.1
|
placenta-specific 8, tandem duplicate 1 |
| chr4_+_3980247 | 0.26 |
ENSDART00000049194
|
gpr37b
|
G protein-coupled receptor 37b |
| chr4_-_5652030 | 0.26 |
ENSDART00000010903
|
rsph9
|
radial spoke head 9 homolog |
| chr12_+_22580579 | 0.26 |
ENSDART00000171725
ENSDART00000192290 |
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr14_-_30587814 | 0.26 |
ENSDART00000144912
ENSDART00000149714 |
tmem265
|
transmembrane protein 265 |
| chr15_-_9272328 | 0.25 |
ENSDART00000172114
|
calm2a
|
calmodulin 2a (phosphorylase kinase, delta) |
| chr12_+_41697664 | 0.25 |
ENSDART00000162302
|
bnip3
|
BCL2 interacting protein 3 |
| chr4_+_12612723 | 0.24 |
ENSDART00000133767
|
lmo3
|
LIM domain only 3 |
| chr2_+_20604775 | 0.24 |
ENSDART00000131501
|
olfml2bb
|
olfactomedin-like 2Bb |
| chr22_+_27090136 | 0.24 |
ENSDART00000136770
|
si:dkey-246e1.3
|
si:dkey-246e1.3 |
| chr24_-_35767501 | 0.24 |
ENSDART00000105680
ENSDART00000042290 ENSDART00000166264 |
dtna
|
dystrobrevin, alpha |
| chr6_+_9870192 | 0.24 |
ENSDART00000150894
|
MPP4 (1 of many)
|
si:ch211-222n4.6 |
| chr4_+_9400012 | 0.23 |
ENSDART00000191960
|
tmtc1
|
transmembrane and tetratricopeptide repeat containing 1 |
| chr8_-_12867434 | 0.23 |
ENSDART00000081657
|
slc2a6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr16_-_51888952 | 0.23 |
ENSDART00000186407
ENSDART00000175435 |
rpl28
|
ribosomal protein L28 |
| chr11_-_41853874 | 0.23 |
ENSDART00000002556
|
mrto4
|
MRT4 homolog, ribosome maturation factor |
| chr10_+_36178713 | 0.23 |
ENSDART00000140816
|
or108-3
|
odorant receptor, family D, subfamily 108, member 3 |
| chr13_+_49175947 | 0.23 |
ENSDART00000056927
|
egln1a
|
egl-9 family hypoxia-inducible factor 1a |
| chr3_-_58798377 | 0.23 |
ENSDART00000161248
|
si:ch73-281f12.4
|
si:ch73-281f12.4 |
| chr3_+_23092762 | 0.22 |
ENSDART00000142884
ENSDART00000024136 |
gngt2a
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2a |
| chr14_+_5385855 | 0.22 |
ENSDART00000031508
|
lbx2
|
ladybird homeobox 2 |
| chr25_-_13842618 | 0.22 |
ENSDART00000160258
|
mapk8ip1a
|
mitogen-activated protein kinase 8 interacting protein 1a |
| chr21_+_15824182 | 0.22 |
ENSDART00000065779
|
gnrh2
|
gonadotropin-releasing hormone 2 |
| chr13_+_22476742 | 0.21 |
ENSDART00000078759
ENSDART00000130101 ENSDART00000137220 ENSDART00000133065 ENSDART00000147348 |
ldb3a
|
LIM domain binding 3a |
| chr21_-_18993110 | 0.21 |
ENSDART00000144086
|
si:ch211-222n4.6
|
si:ch211-222n4.6 |
| chr9_-_20372977 | 0.21 |
ENSDART00000113418
|
igsf3
|
immunoglobulin superfamily, member 3 |
| chr22_-_19552796 | 0.21 |
ENSDART00000148088
ENSDART00000105485 |
si:dkey-78l4.14
|
si:dkey-78l4.14 |
| chr16_-_22930925 | 0.21 |
ENSDART00000133819
|
si:dkey-246i14.3
|
si:dkey-246i14.3 |
| chr1_-_5455498 | 0.21 |
ENSDART00000040368
ENSDART00000114035 |
mnx2b
|
motor neuron and pancreas homeobox 2b |
| chrM_+_8130 | 0.20 |
ENSDART00000093609
|
mt-co2
|
cytochrome c oxidase II, mitochondrial |
| chr3_+_17537352 | 0.20 |
ENSDART00000104549
|
hcrt
|
hypocretin (orexin) neuropeptide precursor |
| chr4_+_72723304 | 0.20 |
ENSDART00000186791
ENSDART00000158902 ENSDART00000191925 |
rab3ip
|
RAB3A interacting protein (rabin3) |
| chr20_-_28642061 | 0.19 |
ENSDART00000135513
|
rgs6
|
regulator of G protein signaling 6 |
| chr8_-_23776399 | 0.19 |
ENSDART00000114800
|
INAVA
|
si:ch211-163l21.4 |
| chr18_+_28106139 | 0.19 |
ENSDART00000089615
|
kiaa1549lb
|
KIAA1549-like b |
| chr1_+_21731382 | 0.19 |
ENSDART00000054395
|
pax5
|
paired box 5 |
| chr15_-_4528326 | 0.19 |
ENSDART00000158122
ENSDART00000155619 ENSDART00000128602 |
tfdp2
|
transcription factor Dp-2 |
| chr2_-_37140423 | 0.19 |
ENSDART00000144220
|
tspan37
|
tetraspanin 37 |
| chr7_+_36898850 | 0.19 |
ENSDART00000113342
|
tox3
|
TOX high mobility group box family member 3 |
| chr2_+_40294313 | 0.18 |
ENSDART00000037292
|
epha4b
|
eph receptor A4b |
| chr17_+_30545895 | 0.18 |
ENSDART00000076739
|
nhsl1a
|
NHS-like 1a |
| chr6_-_55585423 | 0.18 |
ENSDART00000157129
|
slc12a5a
|
solute carrier family 12 (potassium/chloride transporter), member 5a |
| chr1_-_44701313 | 0.18 |
ENSDART00000193926
|
si:dkey-28b4.8
|
si:dkey-28b4.8 |
| chr11_-_40728380 | 0.18 |
ENSDART00000023745
|
ccdc114
|
coiled-coil domain containing 114 |
| chr13_+_49175624 | 0.18 |
ENSDART00000193550
|
egln1a
|
egl-9 family hypoxia-inducible factor 1a |
| chr11_+_28476298 | 0.18 |
ENSDART00000122319
|
lrrc38b
|
leucine rich repeat containing 38b |
| chr23_-_42752550 | 0.18 |
ENSDART00000187059
|
si:ch73-217n20.1
|
si:ch73-217n20.1 |
| chr3_-_29941357 | 0.18 |
ENSDART00000147732
ENSDART00000137973 ENSDART00000103523 |
grna
|
granulin a |
| chr7_-_31759394 | 0.17 |
ENSDART00000193040
|
igdcc4
|
immunoglobulin superfamily, DCC subclass, member 4 |
| chr3_-_58798815 | 0.17 |
ENSDART00000082920
|
si:ch73-281f12.4
|
si:ch73-281f12.4 |
| chr17_+_11675362 | 0.17 |
ENSDART00000157911
|
kif26ba
|
kinesin family member 26Ba |
| chr8_-_25034411 | 0.17 |
ENSDART00000135973
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr17_+_13007307 | 0.17 |
ENSDART00000064554
|
ppp2r3c
|
protein phosphatase 2, regulatory subunit B'', gamma |
| chr10_-_5581487 | 0.17 |
ENSDART00000141943
|
syk
|
spleen tyrosine kinase |
| chr23_-_19230627 | 0.16 |
ENSDART00000007122
|
guca1b
|
guanylate cyclase activator 1B |
| chr21_-_26490186 | 0.16 |
ENSDART00000009889
|
zgc:110540
|
zgc:110540 |
| chr12_-_46112892 | 0.16 |
ENSDART00000187128
ENSDART00000114268 |
zgc:153932
|
zgc:153932 |
| chr8_+_49778486 | 0.16 |
ENSDART00000131732
|
ntrk2a
|
neurotrophic tyrosine kinase, receptor, type 2a |
| chr4_+_4849789 | 0.16 |
ENSDART00000130818
ENSDART00000127751 |
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr16_-_30932533 | 0.16 |
ENSDART00000085761
|
slc45a4
|
solute carrier family 45, member 4 |
| chr10_+_16584382 | 0.16 |
ENSDART00000112039
|
CR790388.1
|
|
| chr14_-_30967284 | 0.16 |
ENSDART00000149435
|
il2rgb
|
interleukin 2 receptor, gamma b |
| chr20_-_48604199 | 0.16 |
ENSDART00000161762
ENSDART00000170894 |
mgst3a
|
microsomal glutathione S-transferase 3a |
| chr11_-_45138857 | 0.16 |
ENSDART00000166501
|
cant1b
|
calcium activated nucleotidase 1b |
| chr2_-_5505094 | 0.16 |
ENSDART00000145035
|
saga
|
S-antigen; retina and pineal gland (arrestin) a |
Network of associatons between targets according to the STRING database.
First level regulatory network of lhx6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) symmetric cell division(GO:0098725) |
| 0.2 | 1.8 | GO:1902868 | positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) |
| 0.2 | 0.6 | GO:0038158 | granulocyte colony-stimulating factor signaling pathway(GO:0038158) |
| 0.2 | 1.0 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.2 | 0.5 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.2 | 0.6 | GO:0014856 | skeletal muscle cell proliferation(GO:0014856) regulation of skeletal muscle cell proliferation(GO:0014857) |
| 0.2 | 0.5 | GO:0008292 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.1 | 1.0 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.1 | 0.6 | GO:0099543 | trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.1 | 1.2 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.1 | 0.4 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 0.7 | GO:0021767 | mammillary body development(GO:0021767) |
| 0.1 | 0.3 | GO:0045579 | positive regulation of B cell differentiation(GO:0045579) |
| 0.1 | 0.9 | GO:0060114 | vestibular receptor cell differentiation(GO:0060114) vestibular receptor cell development(GO:0060118) |
| 0.1 | 0.3 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.1 | 0.3 | GO:0099540 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.1 | 0.3 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.1 | 0.6 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.1 | 0.3 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.1 | 0.6 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.1 | 0.5 | GO:0031498 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.1 | 0.5 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.1 | 0.6 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.1 | 0.2 | GO:2000434 | regulation of protein neddylation(GO:2000434) |
| 0.1 | 0.3 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.2 | GO:0042755 | eating behavior(GO:0042755) |
| 0.0 | 0.4 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.9 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.3 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.5 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.0 | 0.2 | GO:1904105 | positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.0 | 0.3 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.7 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.2 | GO:0097032 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.9 | GO:0014855 | striated muscle cell proliferation(GO:0014855) |
| 0.0 | 0.6 | GO:0030878 | thyroid gland development(GO:0030878) face morphogenesis(GO:0060325) |
| 0.0 | 0.2 | GO:0045046 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.9 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.5 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.4 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.0 | 0.1 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.4 | GO:0046902 | regulation of mitochondrial membrane permeability(GO:0046902) |
| 0.0 | 0.1 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.0 | 0.1 | GO:0071423 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.0 | 0.1 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.1 | GO:0097237 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.0 | 0.6 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.0 | 0.9 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.0 | 0.4 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.0 | 0.1 | GO:0055107 | Golgi to secretory granule transport(GO:0055107) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:0070317 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.7 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.2 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.0 | 0.1 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
| 0.0 | 0.2 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.0 | 0.4 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 0.2 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.6 | GO:0007099 | centriole replication(GO:0007099) |
| 0.0 | 0.5 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.4 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.4 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 0.0 | GO:1903441 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to ciliary membrane(GO:1903441) |
| 0.0 | 0.1 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.0 | 0.3 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.4 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.4 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.1 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.0 | 0.3 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.1 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.0 | 0.2 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.0 | 0.3 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.4 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.0 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.0 | 0.3 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.1 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.3 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.6 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 0.4 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.4 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.1 | 0.7 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.3 | GO:0001534 | radial spoke(GO:0001534) |
| 0.1 | 0.5 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.1 | 0.5 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.6 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.2 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.0 | 0.5 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.0 | 0.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 1.1 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.4 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.3 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.1 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.5 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.3 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.5 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.5 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.2 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.2 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.4 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.3 | 0.8 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.2 | 0.5 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 0.5 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.1 | 0.7 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.3 | GO:0060175 | brain-derived neurotrophic factor binding(GO:0048403) brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.1 | 0.4 | GO:0017064 | fatty acid amide hydrolase activity(GO:0017064) |
| 0.1 | 0.4 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.3 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.2 | GO:0005183 | gonadotropin hormone-releasing hormone activity(GO:0005183) gonadotropin-releasing hormone receptor binding(GO:0031530) |
| 0.1 | 0.3 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.1 | 0.6 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 0.4 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 0.3 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 0.6 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 1.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.2 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 0.6 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.6 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.5 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.2 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.5 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.6 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.1 | GO:1904121 | phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.0 | 0.1 | GO:0015117 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.7 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.5 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.5 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 0.2 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.3 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.2 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.4 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.4 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 1.2 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.7 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.4 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.5 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.0 | 0.3 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.2 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.3 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.0 | 0.0 | GO:0072545 | tyrosine binding(GO:0072545) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.2 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.1 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.7 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 1.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.2 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.4 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.3 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.5 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.5 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.6 | PID NOTCH PATHWAY | Notch signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 0.6 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 0.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.4 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.5 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.6 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.1 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.1 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |