Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
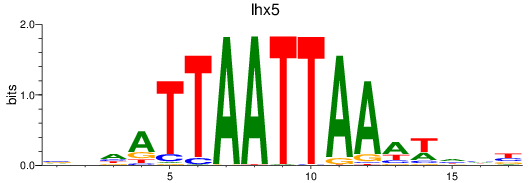
Results for lhx5
Z-value: 0.44
Transcription factors associated with lhx5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
lhx5
|
ENSDARG00000057936 | LIM homeobox 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| lhx5 | dr11_v1_chr21_-_15929041_15929041 | -0.76 | 1.6e-02 | Click! |
Activity profile of lhx5 motif
Sorted Z-values of lhx5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_34916208 | 0.87 |
ENSDART00000187371
|
ccna1
|
cyclin A1 |
| chr15_-_44601331 | 0.77 |
ENSDART00000161514
|
zgc:165508
|
zgc:165508 |
| chr11_-_44801968 | 0.76 |
ENSDART00000161846
|
map1lc3c
|
microtubule-associated protein 1 light chain 3 gamma |
| chr17_-_4245902 | 0.69 |
ENSDART00000151851
|
gdf3
|
growth differentiation factor 3 |
| chr6_-_12912606 | 0.62 |
ENSDART00000164640
|
ical1
|
islet cell autoantigen 1-like |
| chr5_-_48664522 | 0.62 |
ENSDART00000083229
|
mblac2
|
metallo-beta-lactamase domain containing 2 |
| chr19_-_10330778 | 0.54 |
ENSDART00000081465
ENSDART00000136653 ENSDART00000171232 |
ccdc106b
|
coiled-coil domain containing 106b |
| chr25_-_9805269 | 0.53 |
ENSDART00000192048
|
lrrc4c
|
leucine rich repeat containing 4C |
| chr16_+_13965923 | 0.51 |
ENSDART00000103857
|
zgc:162509
|
zgc:162509 |
| chr7_+_24573721 | 0.49 |
ENSDART00000173938
ENSDART00000173681 |
si:dkeyp-75h12.7
|
si:dkeyp-75h12.7 |
| chr14_-_48765262 | 0.49 |
ENSDART00000166463
|
cnot6b
|
CCR4-NOT transcription complex, subunit 6b |
| chr9_+_54039006 | 0.49 |
ENSDART00000112441
|
tlr7
|
toll-like receptor 7 |
| chr18_+_35128685 | 0.48 |
ENSDART00000151579
|
si:ch211-195m9.3
|
si:ch211-195m9.3 |
| chr21_-_2814709 | 0.48 |
ENSDART00000097664
|
SEMA4D
|
semaphorin 4D |
| chr23_-_18707418 | 0.46 |
ENSDART00000144668
ENSDART00000141205 ENSDART00000016765 |
zgc:103759
|
zgc:103759 |
| chr7_+_59212666 | 0.44 |
ENSDART00000172046
|
dok1b
|
docking protein 1b |
| chr1_+_35985813 | 0.44 |
ENSDART00000179634
ENSDART00000139636 ENSDART00000175902 |
zgc:152968
|
zgc:152968 |
| chr17_-_40956035 | 0.44 |
ENSDART00000124715
|
si:dkey-16j16.4
|
si:dkey-16j16.4 |
| chr11_-_26832685 | 0.44 |
ENSDART00000153519
|
iqsec1b
|
IQ motif and Sec7 domain 1b |
| chr12_-_6880694 | 0.43 |
ENSDART00000171846
|
pcdh15b
|
protocadherin-related 15b |
| chr20_+_32552912 | 0.43 |
ENSDART00000009691
|
scml4
|
Scm polycomb group protein like 4 |
| chr4_-_5019113 | 0.41 |
ENSDART00000189321
ENSDART00000081990 |
strip2
|
striatin interacting protein 2 |
| chr6_+_12527725 | 0.41 |
ENSDART00000149328
|
stk24b
|
serine/threonine kinase 24b (STE20 homolog, yeast) |
| chr21_-_44081540 | 0.40 |
ENSDART00000130833
|
FO704810.1
|
|
| chr24_+_17260001 | 0.39 |
ENSDART00000066765
|
bmi1a
|
bmi1 polycomb ring finger oncogene 1a |
| chr3_+_7808459 | 0.39 |
ENSDART00000162374
|
hook2
|
hook microtubule-tethering protein 2 |
| chr9_-_27398369 | 0.39 |
ENSDART00000186499
|
tex30
|
testis expressed 30 |
| chr24_+_17260329 | 0.39 |
ENSDART00000129554
|
bmi1a
|
bmi1 polycomb ring finger oncogene 1a |
| chr17_-_25831569 | 0.38 |
ENSDART00000148743
|
hhat
|
hedgehog acyltransferase |
| chr13_+_35528607 | 0.37 |
ENSDART00000075414
ENSDART00000112947 |
wdr27
|
WD repeat domain 27 |
| chr14_-_32631519 | 0.37 |
ENSDART00000167282
ENSDART00000052938 |
atp11c
|
ATPase phospholipid transporting 11C |
| chr8_+_36500308 | 0.36 |
ENSDART00000098701
|
slc7a4
|
solute carrier family 7, member 4 |
| chr23_-_18024543 | 0.36 |
ENSDART00000139695
|
pm20d1.1
|
peptidase M20 domain containing 1, tandem duplicate 1 |
| chr14_-_32631013 | 0.36 |
ENSDART00000176815
|
atp11c
|
ATPase phospholipid transporting 11C |
| chr16_-_42965192 | 0.35 |
ENSDART00000113714
|
mtx1a
|
metaxin 1a |
| chr10_-_2524917 | 0.34 |
ENSDART00000188642
|
CU856539.1
|
|
| chr2_-_17392799 | 0.33 |
ENSDART00000136470
ENSDART00000141188 |
st3gal3b
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3b |
| chr8_+_36500061 | 0.33 |
ENSDART00000185840
|
slc7a4
|
solute carrier family 7, member 4 |
| chr10_-_21362071 | 0.33 |
ENSDART00000125167
|
avd
|
avidin |
| chr4_-_73447058 | 0.32 |
ENSDART00000172042
|
si:ch73-120g24.4
|
si:ch73-120g24.4 |
| chr16_+_2820340 | 0.32 |
ENSDART00000092299
ENSDART00000192931 ENSDART00000148512 |
si:dkey-288i20.2
|
si:dkey-288i20.2 |
| chr3_-_11624694 | 0.31 |
ENSDART00000127157
|
hlfa
|
hepatic leukemia factor a |
| chr17_-_27048537 | 0.31 |
ENSDART00000050018
ENSDART00000193861 |
cnksr1
|
connector enhancer of kinase suppressor of Ras 1 |
| chr10_-_21362320 | 0.30 |
ENSDART00000189789
|
avd
|
avidin |
| chr16_-_10223741 | 0.30 |
ENSDART00000188099
|
si:rp71-15i12.1
|
si:rp71-15i12.1 |
| chr8_+_7801060 | 0.30 |
ENSDART00000161618
|
tfe3a
|
transcription factor binding to IGHM enhancer 3a |
| chr6_-_57539141 | 0.29 |
ENSDART00000156967
|
itcha
|
itchy E3 ubiquitin protein ligase a |
| chr2_-_17393216 | 0.29 |
ENSDART00000123137
|
st3gal3b
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3b |
| chr16_-_28658341 | 0.29 |
ENSDART00000148456
|
abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr11_-_34478225 | 0.29 |
ENSDART00000189604
|
xxylt1
|
xyloside xylosyltransferase 1 |
| chr18_-_41161828 | 0.28 |
ENSDART00000114993
|
CABZ01005876.1
|
|
| chr8_-_11834599 | 0.28 |
ENSDART00000190986
|
rapgef1a
|
Rap guanine nucleotide exchange factor (GEF) 1a |
| chr5_-_32396929 | 0.28 |
ENSDART00000023977
|
fbxw2
|
F-box and WD repeat domain containing 2 |
| chr22_-_20812822 | 0.28 |
ENSDART00000193778
|
dot1l
|
DOT1-like histone H3K79 methyltransferase |
| chr10_-_2524297 | 0.28 |
ENSDART00000192475
|
CU856539.1
|
|
| chr3_-_26787430 | 0.27 |
ENSDART00000087047
|
rab40c
|
RAB40c, member RAS oncogene family |
| chr22_-_12304591 | 0.27 |
ENSDART00000136408
|
zranb3
|
zinc finger, RAN-binding domain containing 3 |
| chr21_-_20939488 | 0.27 |
ENSDART00000039043
|
rgs7bpb
|
regulator of G protein signaling 7 binding protein b |
| chr9_+_48761455 | 0.27 |
ENSDART00000139631
|
abcb11a
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11a |
| chr25_-_27621268 | 0.27 |
ENSDART00000146205
ENSDART00000073511 |
hyal6
|
hyaluronoglucosaminidase 6 |
| chr4_-_75158035 | 0.27 |
ENSDART00000174353
|
CABZ01066312.1
|
|
| chr6_+_49551614 | 0.26 |
ENSDART00000022581
|
rab22a
|
RAB22A, member RAS oncogene family |
| chr10_-_31015535 | 0.26 |
ENSDART00000146116
|
panx3
|
pannexin 3 |
| chr21_-_22635245 | 0.25 |
ENSDART00000115224
ENSDART00000101782 |
nectin1a
|
nectin cell adhesion molecule 1a |
| chr21_+_21195487 | 0.25 |
ENSDART00000181746
ENSDART00000184832 |
rictorb
|
RPTOR independent companion of MTOR, complex 2b |
| chr3_-_48716422 | 0.25 |
ENSDART00000164979
|
si:ch211-114m9.1
|
si:ch211-114m9.1 |
| chr12_-_48188928 | 0.24 |
ENSDART00000184384
|
pald1a
|
phosphatase domain containing, paladin 1a |
| chr4_+_25692277 | 0.24 |
ENSDART00000047113
|
acot18
|
acyl-CoA thioesterase 18 |
| chr9_-_50001606 | 0.24 |
ENSDART00000161648
ENSDART00000168514 |
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr7_-_57509447 | 0.24 |
ENSDART00000051973
ENSDART00000147036 |
sirt3
|
sirtuin 3 |
| chr2_+_42072231 | 0.24 |
ENSDART00000084517
|
vcpip1
|
valosin containing protein (p97)/p47 complex interacting protein 1 |
| chr1_+_47499888 | 0.23 |
ENSDART00000027624
|
stn1
|
STN1, CST complex subunit |
| chr4_+_2482046 | 0.23 |
ENSDART00000103371
|
zdhhc17
|
zinc finger, DHHC-type containing 17 |
| chr2_+_9990491 | 0.23 |
ENSDART00000011906
|
slc35a3b
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3b |
| chr10_-_13343831 | 0.23 |
ENSDART00000135941
|
il11ra
|
interleukin 11 receptor, alpha |
| chr21_+_42717424 | 0.23 |
ENSDART00000166936
ENSDART00000172135 |
sh3pxd2b
|
SH3 and PX domains 2B |
| chr9_+_50001746 | 0.22 |
ENSDART00000058892
|
slc38a11
|
solute carrier family 38, member 11 |
| chr19_+_40069524 | 0.22 |
ENSDART00000151365
ENSDART00000140926 |
zmym4
|
zinc finger, MYM-type 4 |
| chr2_+_10878406 | 0.22 |
ENSDART00000091497
|
tceanc2
|
transcription elongation factor A (SII) N-terminal and central domain containing 2 |
| chr22_+_737211 | 0.22 |
ENSDART00000017305
|
znf76
|
zinc finger protein 76 |
| chr8_-_25569920 | 0.21 |
ENSDART00000136869
|
prex1
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
| chr19_+_2631565 | 0.21 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr17_+_43595692 | 0.21 |
ENSDART00000156271
|
cfap99
|
cilia and flagella associated protein 99 |
| chr10_-_1276046 | 0.21 |
ENSDART00000169779
|
pdlim5b
|
PDZ and LIM domain 5b |
| chr25_+_22320738 | 0.21 |
ENSDART00000073566
|
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr1_+_418869 | 0.21 |
ENSDART00000152173
|
tpp2
|
tripeptidyl peptidase 2 |
| chr20_+_22799857 | 0.21 |
ENSDART00000058527
|
scfd2
|
sec1 family domain containing 2 |
| chr7_+_66884291 | 0.21 |
ENSDART00000187499
|
sbf2
|
SET binding factor 2 |
| chr15_+_34592215 | 0.20 |
ENSDART00000099776
|
tspan13a
|
tetraspanin 13a |
| chr15_-_36727462 | 0.20 |
ENSDART00000085971
|
nphs1
|
nephrosis 1, congenital, Finnish type (nephrin) |
| chr17_-_16422654 | 0.20 |
ENSDART00000150149
|
tdp1
|
tyrosyl-DNA phosphodiesterase 1 |
| chr9_-_37749973 | 0.20 |
ENSDART00000087663
|
sema5ba
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5Ba |
| chr17_+_28533102 | 0.19 |
ENSDART00000156218
|
mdga2a
|
MAM domain containing glycosylphosphatidylinositol anchor 2a |
| chr10_+_16225117 | 0.19 |
ENSDART00000169885
|
slc12a2
|
solute carrier family 12 (sodium/potassium/chloride transporter), member 2 |
| chr22_-_14739491 | 0.19 |
ENSDART00000133385
|
lrp1ba
|
low density lipoprotein receptor-related protein 1Ba |
| chr23_-_19686791 | 0.19 |
ENSDART00000161973
|
zgc:193598
|
zgc:193598 |
| chr25_-_25058508 | 0.19 |
ENSDART00000087570
ENSDART00000178891 |
FQ311928.1
|
|
| chr22_+_30047245 | 0.19 |
ENSDART00000142857
ENSDART00000141247 ENSDART00000140015 ENSDART00000040538 |
add3a
|
adducin 3 (gamma) a |
| chr23_+_16638639 | 0.19 |
ENSDART00000143545
|
snphb
|
syntaphilin b |
| chr9_+_7998794 | 0.18 |
ENSDART00000138167
|
myo16
|
myosin XVI |
| chr4_-_2219705 | 0.18 |
ENSDART00000131046
|
si:ch73-278m9.1
|
si:ch73-278m9.1 |
| chr8_+_26007308 | 0.18 |
ENSDART00000191423
|
xpc
|
xeroderma pigmentosum, complementation group C |
| chr20_+_22799641 | 0.18 |
ENSDART00000131132
|
scfd2
|
sec1 family domain containing 2 |
| chr25_-_37186894 | 0.18 |
ENSDART00000191647
ENSDART00000182095 |
tdrd12
|
tudor domain containing 12 |
| chr25_-_21031007 | 0.18 |
ENSDART00000138985
|
gnaia
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a |
| chr25_-_13490744 | 0.18 |
ENSDART00000056721
|
ldhd
|
lactate dehydrogenase D |
| chr23_-_17509656 | 0.18 |
ENSDART00000148423
|
dnajc5ab
|
DnaJ (Hsp40) homolog, subfamily C, member 5ab |
| chr5_-_21422390 | 0.18 |
ENSDART00000144198
|
tenm1
|
teneurin transmembrane protein 1 |
| chr13_+_11440389 | 0.17 |
ENSDART00000186463
|
zbtb18
|
zinc finger and BTB domain containing 18 |
| chr1_-_51038885 | 0.17 |
ENSDART00000035150
|
spast
|
spastin |
| chr22_-_13466246 | 0.17 |
ENSDART00000134035
|
cntnap5b
|
contactin associated protein-like 5b |
| chr11_-_44979281 | 0.16 |
ENSDART00000190972
|
ldb1b
|
LIM-domain binding 1b |
| chr2_-_10877765 | 0.16 |
ENSDART00000100607
|
cdc7
|
cell division cycle 7 homolog (S. cerevisiae) |
| chr9_-_38398789 | 0.16 |
ENSDART00000188384
|
znf142
|
zinc finger protein 142 |
| chr13_-_38730267 | 0.16 |
ENSDART00000157524
|
lmbrd1
|
LMBR1 domain containing 1 |
| chr21_+_45502773 | 0.16 |
ENSDART00000160059
ENSDART00000165704 |
si:dkey-223p19.2
|
si:dkey-223p19.2 |
| chr16_-_31622777 | 0.16 |
ENSDART00000137311
ENSDART00000002930 |
phf20l1
|
PHD finger protein 20 like 1 |
| chr7_+_66884570 | 0.16 |
ENSDART00000082664
|
sbf2
|
SET binding factor 2 |
| chr7_+_21272833 | 0.15 |
ENSDART00000052942
|
serpinh2
|
serine (or cysteine) peptidase inhibitor, clade H, member 2 |
| chr4_-_5018705 | 0.15 |
ENSDART00000154025
|
strip2
|
striatin interacting protein 2 |
| chr2_-_16217344 | 0.15 |
ENSDART00000152031
|
arhgef4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr9_+_24065855 | 0.15 |
ENSDART00000161468
ENSDART00000171577 ENSDART00000172743 ENSDART00000159324 ENSDART00000079689 ENSDART00000023196 ENSDART00000101577 |
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr12_+_45200744 | 0.15 |
ENSDART00000098932
|
wbp2
|
WW domain binding protein 2 |
| chr1_-_9195629 | 0.15 |
ENSDART00000143587
ENSDART00000192174 |
ern2
|
endoplasmic reticulum to nucleus signaling 2 |
| chr13_+_33268657 | 0.14 |
ENSDART00000002095
|
tmem39b
|
transmembrane protein 39B |
| chr7_-_71531846 | 0.14 |
ENSDART00000111797
|
acer2
|
alkaline ceramidase 2 |
| chr23_-_31969786 | 0.14 |
ENSDART00000134550
|
ormdl2
|
ORMDL sphingolipid biosynthesis regulator 2 |
| chr6_-_50730749 | 0.13 |
ENSDART00000157153
ENSDART00000110441 |
pigu
|
phosphatidylinositol glycan anchor biosynthesis, class U |
| chr1_-_45616470 | 0.13 |
ENSDART00000150165
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr14_+_44794936 | 0.13 |
ENSDART00000128881
|
zgc:195212
|
zgc:195212 |
| chr1_+_41498188 | 0.13 |
ENSDART00000191934
ENSDART00000146310 |
dtx4
|
deltex 4, E3 ubiquitin ligase |
| chr23_+_4709607 | 0.13 |
ENSDART00000166503
ENSDART00000158752 ENSDART00000163860 ENSDART00000172739 |
raf1a
raf1a
|
Raf-1 proto-oncogene, serine/threonine kinase a Raf-1 proto-oncogene, serine/threonine kinase a |
| chr17_-_200316 | 0.13 |
ENSDART00000190561
|
CABZ01083778.1
|
|
| chr6_-_1865323 | 0.13 |
ENSDART00000155775
|
dlgap4b
|
discs, large (Drosophila) homolog-associated protein 4b |
| chr6_+_40922572 | 0.13 |
ENSDART00000133599
ENSDART00000002728 ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr7_+_20383841 | 0.13 |
ENSDART00000052906
|
si:dkey-33c9.6
|
si:dkey-33c9.6 |
| chr4_-_5108844 | 0.13 |
ENSDART00000132666
ENSDART00000136096 |
tmem209
|
transmembrane protein 209 |
| chr21_+_43172506 | 0.12 |
ENSDART00000121725
|
zcchc10
|
zinc finger, CCHC domain containing 10 |
| chr2_-_9059955 | 0.12 |
ENSDART00000022768
|
ak5
|
adenylate kinase 5 |
| chr5_+_32076109 | 0.12 |
ENSDART00000051357
ENSDART00000144510 |
zmat5
|
zinc finger, matrin-type 5 |
| chr15_-_20949692 | 0.12 |
ENSDART00000185548
|
tbcela
|
tubulin folding cofactor E-like a |
| chr24_-_25166720 | 0.12 |
ENSDART00000141601
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr4_+_77943184 | 0.12 |
ENSDART00000159094
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr22_-_28226948 | 0.11 |
ENSDART00000147686
|
si:dkey-222p3.1
|
si:dkey-222p3.1 |
| chr7_-_74057288 | 0.11 |
ENSDART00000189740
|
CU929544.1
|
|
| chr19_+_46158078 | 0.11 |
ENSDART00000183933
ENSDART00000164055 |
cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr5_+_69747417 | 0.11 |
ENSDART00000153717
|
si:ch211-275j6.5
|
si:ch211-275j6.5 |
| chr25_-_1126311 | 0.11 |
ENSDART00000161863
|
spg11
|
spastic paraplegia 11 |
| chr23_+_1029450 | 0.11 |
ENSDART00000189196
|
si:zfos-905g2.1
|
si:zfos-905g2.1 |
| chr8_+_42718135 | 0.11 |
ENSDART00000158900
|
zgc:194007
|
zgc:194007 |
| chr25_+_3994823 | 0.11 |
ENSDART00000154020
|
eps8l2
|
EPS8 like 2 |
| chr3_-_26806032 | 0.11 |
ENSDART00000143710
|
pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr14_+_16151368 | 0.11 |
ENSDART00000160973
|
polr1a
|
polymerase (RNA) I polypeptide A |
| chr14_-_16151055 | 0.10 |
ENSDART00000162431
|
ptcd3
|
pentatricopeptide repeat domain 3 |
| chr19_+_5480327 | 0.10 |
ENSDART00000148794
|
jupb
|
junction plakoglobin b |
| chr10_-_43721530 | 0.10 |
ENSDART00000025366
|
cetn3
|
centrin 3 |
| chr24_+_26329018 | 0.10 |
ENSDART00000145752
|
mynn
|
myoneurin |
| chr21_-_25801956 | 0.10 |
ENSDART00000101219
|
mettl27
|
methyltransferase like 27 |
| chr18_+_15644559 | 0.10 |
ENSDART00000061794
|
nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr23_-_2901167 | 0.10 |
ENSDART00000165955
ENSDART00000190616 |
zhx3
|
zinc fingers and homeoboxes 3 |
| chr11_+_17984354 | 0.10 |
ENSDART00000179986
|
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr9_+_7732714 | 0.10 |
ENSDART00000145853
|
si:ch1073-349o24.2
|
si:ch1073-349o24.2 |
| chr9_-_9415000 | 0.09 |
ENSDART00000146210
|
si:ch211-214p13.9
|
si:ch211-214p13.9 |
| chr14_+_16151636 | 0.09 |
ENSDART00000159352
|
polr1a
|
polymerase (RNA) I polypeptide A |
| chr3_+_40164129 | 0.09 |
ENSDART00000102526
|
gfer
|
growth factor, augmenter of liver regeneration (ERV1 homolog, S. cerevisiae) |
| chr1_+_32054159 | 0.09 |
ENSDART00000181442
|
sts
|
steroid sulfatase (microsomal), isozyme S |
| chr1_-_50438247 | 0.09 |
ENSDART00000114098
|
dkk2
|
dickkopf WNT signaling pathway inhibitor 2 |
| chr16_+_24733741 | 0.09 |
ENSDART00000155217
|
si:dkey-79d12.4
|
si:dkey-79d12.4 |
| chr13_+_9468535 | 0.09 |
ENSDART00000135088
ENSDART00000164270 ENSDART00000099619 ENSDART00000164656 |
HTRA2 (1 of many)
|
si:dkey-265c15.6 |
| chr23_-_18567088 | 0.09 |
ENSDART00000192371
|
sephs2
|
selenophosphate synthetase 2 |
| chr24_+_26328787 | 0.09 |
ENSDART00000003884
|
mynn
|
myoneurin |
| chr10_-_11261565 | 0.09 |
ENSDART00000146727
|
ptbp3
|
polypyrimidine tract binding protein 3 |
| chr22_+_30046365 | 0.09 |
ENSDART00000192546
|
add3a
|
adducin 3 (gamma) a |
| chr5_-_23843636 | 0.09 |
ENSDART00000193280
|
GBGT1 (1 of many)
|
si:ch211-135f11.5 |
| chr11_+_45436703 | 0.09 |
ENSDART00000168295
ENSDART00000173293 |
sos1
|
son of sevenless homolog 1 (Drosophila) |
| chr13_-_29421331 | 0.08 |
ENSDART00000150228
|
chata
|
choline O-acetyltransferase a |
| chr18_+_8320165 | 0.08 |
ENSDART00000092053
|
chkb
|
choline kinase beta |
| chr9_+_38074082 | 0.08 |
ENSDART00000017833
|
cacnb4a
|
calcium channel, voltage-dependent, beta 4a subunit |
| chr20_+_25586099 | 0.08 |
ENSDART00000063122
ENSDART00000134047 |
cyp2p10
|
cytochrome P450, family 2, subfamily P, polypeptide 10 |
| chr4_+_20536120 | 0.08 |
ENSDART00000125685
|
zmp:0000000650
|
zmp:0000000650 |
| chr3_+_39853788 | 0.08 |
ENSDART00000154869
|
cacna1ha
|
calcium channel, voltage-dependent, T type, alpha 1H subunit a |
| chr8_+_50953776 | 0.08 |
ENSDART00000013870
|
zgc:56596
|
zgc:56596 |
| chr22_+_2751887 | 0.08 |
ENSDART00000133652
|
si:dkey-20i20.11
|
si:dkey-20i20.11 |
| chr7_-_7692723 | 0.08 |
ENSDART00000183352
|
aadat
|
aminoadipate aminotransferase |
| chr22_-_11614973 | 0.08 |
ENSDART00000063135
|
pcyt2
|
phosphate cytidylyltransferase 2, ethanolamine |
| chr9_-_43644261 | 0.08 |
ENSDART00000023684
|
cwc22
|
CWC22 spliceosome-associated protein homolog (S. cerevisiae) |
| chr1_+_23563691 | 0.08 |
ENSDART00000142879
|
ncapg
|
non-SMC condensin I complex, subunit G |
| chr1_+_54124209 | 0.07 |
ENSDART00000187730
|
LO017722.1
|
|
| chr11_+_17984167 | 0.07 |
ENSDART00000020283
ENSDART00000188329 |
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr3_+_23029484 | 0.07 |
ENSDART00000187900
|
nags
|
N-acetylglutamate synthase |
| chr2_+_11923615 | 0.07 |
ENSDART00000126118
|
trove2
|
TROVE domain family, member 2 |
| chr23_+_39970425 | 0.07 |
ENSDART00000165376
|
fyco1a
|
FYVE and coiled-coil domain containing 1a |
| chr9_+_54290896 | 0.07 |
ENSDART00000149175
|
pou4f3
|
POU class 4 homeobox 3 |
| chr8_-_14239023 | 0.07 |
ENSDART00000090371
|
sin3b
|
SIN3 transcription regulator family member B |
| chr8_+_36560019 | 0.07 |
ENSDART00000136418
ENSDART00000061378 ENSDART00000185237 |
sf3a1
|
splicing factor 3a, subunit 1 |
| chr3_-_7546740 | 0.07 |
ENSDART00000128960
|
zmp:0000001003
|
zmp:0000001003 |
| chr16_-_41004731 | 0.07 |
ENSDART00000102591
|
KCNV1
|
si:dkey-201i6.2 |
| chr4_-_948776 | 0.06 |
ENSDART00000023483
|
sim1b
|
single-minded family bHLH transcription factor 1b |
| chr5_+_21144269 | 0.06 |
ENSDART00000028087
|
cds2
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 2 |
| chr10_-_25716841 | 0.06 |
ENSDART00000140483
ENSDART00000125665 ENSDART00000181678 |
scaf4a
|
SR-related CTD-associated factor 4a |
Network of associatons between targets according to the STRING database.
First level regulatory network of lhx5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.1 | 0.7 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.1 | 0.4 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.1 | 0.3 | GO:0019062 | virion attachment to host cell(GO:0019062) adhesion of symbiont to host(GO:0044406) adhesion of symbiont to host cell(GO:0044650) |
| 0.1 | 0.3 | GO:0034729 | histone H3-K79 methylation(GO:0034729) regulation of transcription regulatory region DNA binding(GO:2000677) |
| 0.1 | 0.3 | GO:0015722 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.1 | 0.2 | GO:0021611 | facial nerve formation(GO:0021611) |
| 0.1 | 0.2 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.1 | 0.2 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.1 | 0.2 | GO:0048313 | organelle inheritance(GO:0048308) Golgi inheritance(GO:0048313) |
| 0.1 | 0.2 | GO:0051228 | protein hexamerization(GO:0034214) mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.1 | 0.8 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.2 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.0 | 0.8 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.3 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.5 | GO:0016074 | snoRNA metabolic process(GO:0016074) |
| 0.0 | 0.2 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.0 | 0.6 | GO:0055117 | regulation of cardiac muscle contraction(GO:0055117) |
| 0.0 | 0.2 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.0 | 0.2 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.0 | 0.3 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.0 | 0.4 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.1 | GO:0090155 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.0 | 0.3 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.0 | 0.1 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.0 | 0.2 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.0 | 0.1 | GO:1900620 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.0 | 0.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.7 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.1 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.3 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.3 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.2 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.0 | 0.2 | GO:0048796 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.1 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.9 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.1 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.2 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.2 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.1 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.5 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.3 | GO:0016233 | telomere capping(GO:0016233) |
| 0.0 | 0.1 | GO:0001765 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.1 | GO:0040033 | miRNA metabolic process(GO:0010586) miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0090309 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.1 | 0.2 | GO:1990879 | CST complex(GO:1990879) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.4 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.8 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.5 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.4 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.8 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.1 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.2 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.2 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.0 | 0.1 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.0 | 0.1 | GO:0071012 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 0.1 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.0 | 0.2 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.3 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.2 | GO:0031932 | TORC2 complex(GO:0031932) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:1990174 | phosphodiesterase decapping endonuclease activity(GO:1990174) |
| 0.1 | 0.6 | GO:0009374 | biotin binding(GO:0009374) |
| 0.1 | 0.3 | GO:0015562 | protein-transmembrane transporting ATPase activity(GO:0015462) efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 0.6 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.1 | 0.2 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.1 | 0.3 | GO:0031151 | histone methyltransferase activity (H3-K79 specific)(GO:0031151) |
| 0.1 | 0.2 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.1 | 0.3 | GO:0015432 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.1 | 0.2 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.2 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.3 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.0 | 0.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.8 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.4 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.2 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0034618 | arginine binding(GO:0034618) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.2 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.2 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.0 | 0.2 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.2 | GO:0098847 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.3 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.2 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.1 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.2 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.1 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.9 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.2 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.2 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.8 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.3 | PID IL3 PATHWAY | IL3-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.5 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |