Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
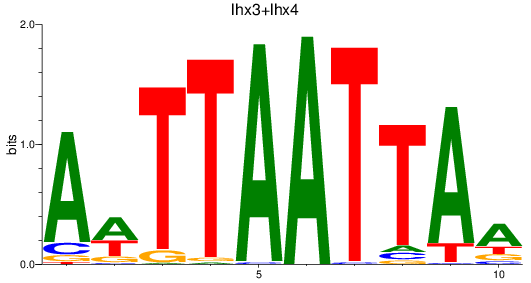
Results for lhx3+lhx4
Z-value: 1.43
Transcription factors associated with lhx3+lhx4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
lhx3
|
ENSDARG00000003803 | LIM homeobox 3 |
|
lhx4
|
ENSDARG00000039458 | LIM homeobox 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| lhx4 | dr11_v1_chr8_-_14484599_14484599 | 0.91 | 6.5e-04 | Click! |
| LHX3 | dr11_v1_chr5_+_71802014_71802014 | 0.87 | 2.2e-03 | Click! |
Activity profile of lhx3+lhx4 motif
Sorted Z-values of lhx3+lhx4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_+_31276842 | 5.09 |
ENSDART00000187238
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr9_+_34641237 | 4.34 |
ENSDART00000133996
|
shox
|
short stature homeobox |
| chr4_+_9669717 | 4.05 |
ENSDART00000004604
|
si:dkey-153k10.9
|
si:dkey-153k10.9 |
| chr9_-_22099536 | 3.79 |
ENSDART00000101923
|
CR391987.1
|
|
| chr3_+_26145013 | 2.99 |
ENSDART00000162546
ENSDART00000129561 |
atp2a1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr17_+_15433518 | 2.98 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr13_+_24279021 | 2.92 |
ENSDART00000058629
|
acta1b
|
actin, alpha 1b, skeletal muscle |
| chr20_-_48485354 | 2.86 |
ENSDART00000124040
ENSDART00000148437 |
insm1a
|
insulinoma-associated 1a |
| chr17_+_15433671 | 2.84 |
ENSDART00000149568
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr14_+_46313396 | 2.51 |
ENSDART00000047525
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr5_-_41494831 | 2.25 |
ENSDART00000051081
|
eef2l2
|
eukaryotic translation elongation factor 2, like 2 |
| chr7_+_29954709 | 2.20 |
ENSDART00000173904
|
tpma
|
alpha-tropomyosin |
| chr14_-_413273 | 2.11 |
ENSDART00000163976
ENSDART00000179907 |
FAT4
|
FAT atypical cadherin 4 |
| chr15_-_23376541 | 2.03 |
ENSDART00000078570
|
c1qtnf5
|
C1q and TNF related 5 |
| chr20_+_19512727 | 2.01 |
ENSDART00000063696
|
atraid
|
all-trans retinoic acid-induced differentiation factor |
| chr12_+_27331324 | 1.98 |
ENSDART00000087208
|
sost
|
sclerostin |
| chr20_+_34933183 | 1.92 |
ENSDART00000062738
|
snap25a
|
synaptosomal-associated protein, 25a |
| chr22_+_16535575 | 1.91 |
ENSDART00000083063
|
tal1
|
T-cell acute lymphocytic leukemia 1 |
| chr5_-_67911111 | 1.90 |
ENSDART00000051833
|
gsx1
|
GS homeobox 1 |
| chr7_-_28148310 | 1.89 |
ENSDART00000044208
|
lmo1
|
LIM domain only 1 |
| chr9_-_42418470 | 1.87 |
ENSDART00000144353
|
calcrla
|
calcitonin receptor-like a |
| chr14_-_26536504 | 1.85 |
ENSDART00000105933
|
tgfbi
|
transforming growth factor, beta-induced |
| chr16_+_33593116 | 1.83 |
ENSDART00000013148
|
pou3f1
|
POU class 3 homeobox 1 |
| chr12_-_35830625 | 1.77 |
ENSDART00000180028
|
CU459056.1
|
|
| chr1_-_40911332 | 1.76 |
ENSDART00000027463
|
hmx4
|
H6 family homeobox 4 |
| chr15_-_15449929 | 1.65 |
ENSDART00000101918
|
proca1
|
protein interacting with cyclin A1 |
| chr10_-_43771447 | 1.64 |
ENSDART00000052307
|
arrdc3b
|
arrestin domain containing 3b |
| chr8_-_34052019 | 1.64 |
ENSDART00000040126
ENSDART00000159208 ENSDART00000048994 ENSDART00000098822 |
pbx3b
|
pre-B-cell leukemia homeobox 3b |
| chr20_+_4060839 | 1.61 |
ENSDART00000178565
|
TRIM67
|
tripartite motif containing 67 |
| chr7_+_31891110 | 1.58 |
ENSDART00000173883
|
mybpc3
|
myosin binding protein C, cardiac |
| chr2_-_30668580 | 1.57 |
ENSDART00000087270
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr19_+_43297546 | 1.56 |
ENSDART00000168002
|
laptm5
|
lysosomal protein transmembrane 5 |
| chr6_-_20952187 | 1.55 |
ENSDART00000074327
|
igfbp2a
|
insulin-like growth factor binding protein 2a |
| chr9_-_14108896 | 1.54 |
ENSDART00000135209
|
prkag3b
|
protein kinase, AMP-activated, gamma 3b non-catalytic subunit |
| chr24_+_24461558 | 1.50 |
ENSDART00000182424
|
bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr16_+_46111849 | 1.50 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr11_+_25257022 | 1.49 |
ENSDART00000156052
|
tp53inp2
|
tumor protein p53 inducible nuclear protein 2 |
| chr24_+_24461341 | 1.48 |
ENSDART00000147658
|
bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr5_+_71802014 | 1.46 |
ENSDART00000124939
ENSDART00000097164 |
LHX3
|
LIM homeobox 3 |
| chr19_+_12915498 | 1.45 |
ENSDART00000132892
|
cthrc1a
|
collagen triple helix repeat containing 1a |
| chr11_-_6048490 | 1.41 |
ENSDART00000066164
|
plvapb
|
plasmalemma vesicle associated protein b |
| chr23_-_39666519 | 1.40 |
ENSDART00000110868
ENSDART00000190961 |
vwa1
|
von Willebrand factor A domain containing 1 |
| chr7_+_26629084 | 1.40 |
ENSDART00000101044
ENSDART00000173765 |
hsbp1a
|
heat shock factor binding protein 1a |
| chr21_+_25236297 | 1.40 |
ENSDART00000112783
|
tmem45b
|
transmembrane protein 45B |
| chr7_+_25033924 | 1.39 |
ENSDART00000170873
|
sb:cb1058
|
sb:cb1058 |
| chr2_+_2223837 | 1.35 |
ENSDART00000101038
ENSDART00000129354 |
tmie
|
transmembrane inner ear |
| chr11_-_1509773 | 1.32 |
ENSDART00000050762
|
phactr3b
|
phosphatase and actin regulator 3b |
| chr3_+_46635527 | 1.31 |
ENSDART00000153971
|
si:dkey-248g21.1
|
si:dkey-248g21.1 |
| chr6_+_23887314 | 1.28 |
ENSDART00000163188
|
znf648
|
zinc finger protein 648 |
| chr17_-_29119362 | 1.27 |
ENSDART00000104204
|
foxg1a
|
forkhead box G1a |
| chr13_-_9886579 | 1.26 |
ENSDART00000101926
|
si:ch211-117n7.7
|
si:ch211-117n7.7 |
| chr11_+_41981959 | 1.26 |
ENSDART00000055707
|
her15.1
|
hairy and enhancer of split-related 15, tandem duplicate 1 |
| chr6_+_52350443 | 1.26 |
ENSDART00000151612
ENSDART00000151349 |
si:ch211-239j9.1
|
si:ch211-239j9.1 |
| chr25_+_31227747 | 1.25 |
ENSDART00000033872
|
tnni2a.1
|
troponin I type 2a (skeletal, fast), tandem duplicate 1 |
| chr9_+_307863 | 1.25 |
ENSDART00000163474
|
stac3
|
SH3 and cysteine rich domain 3 |
| chr21_+_45841731 | 1.23 |
ENSDART00000038657
|
faxdc2
|
fatty acid hydroxylase domain containing 2 |
| chr19_-_5699703 | 1.21 |
ENSDART00000082050
|
zgc:174904
|
zgc:174904 |
| chr16_-_28856112 | 1.20 |
ENSDART00000078543
|
syt11b
|
synaptotagmin XIb |
| chr24_-_26310854 | 1.20 |
ENSDART00000080113
|
apodb
|
apolipoprotein Db |
| chr7_-_58729894 | 1.19 |
ENSDART00000149347
|
chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr17_-_42213285 | 1.17 |
ENSDART00000140549
|
nkx2.2a
|
NK2 homeobox 2a |
| chr3_-_56456527 | 1.16 |
ENSDART00000156553
|
cyth1a
|
cytohesin 1a |
| chr1_+_19535144 | 1.14 |
ENSDART00000103089
|
si:dkey-245p14.4
|
si:dkey-245p14.4 |
| chr1_-_31534089 | 1.14 |
ENSDART00000007770
|
lbx1b
|
ladybird homeobox 1b |
| chr3_+_32553714 | 1.12 |
ENSDART00000165638
|
pax10
|
paired box 10 |
| chr15_-_21014270 | 1.10 |
ENSDART00000154019
|
si:ch211-212c13.10
|
si:ch211-212c13.10 |
| chr21_-_20939488 | 1.10 |
ENSDART00000039043
|
rgs7bpb
|
regulator of G protein signaling 7 binding protein b |
| chr15_+_47440477 | 1.10 |
ENSDART00000002384
|
phox2a
|
paired-like homeobox 2a |
| chr15_-_34567370 | 1.10 |
ENSDART00000099793
|
sostdc1a
|
sclerostin domain containing 1a |
| chr25_+_13406069 | 1.08 |
ENSDART00000010495
|
znrf1
|
zinc and ring finger 1 |
| chr18_+_21408794 | 1.07 |
ENSDART00000140161
|
necab2
|
N-terminal EF-hand calcium binding protein 2 |
| chr1_+_12766351 | 1.06 |
ENSDART00000165785
|
pcdh10a
|
protocadherin 10a |
| chr24_-_38657683 | 1.06 |
ENSDART00000154843
|
si:ch1073-164k15.3
|
si:ch1073-164k15.3 |
| chr1_-_669717 | 1.06 |
ENSDART00000160564
|
cyyr1
|
cysteine/tyrosine-rich 1 |
| chr19_-_4010263 | 1.06 |
ENSDART00000159605
ENSDART00000165541 |
map7d1b
|
MAP7 domain containing 1b |
| chr6_-_11768198 | 1.05 |
ENSDART00000183463
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr1_+_6172786 | 1.04 |
ENSDART00000126468
|
prkag3a
|
protein kinase, AMP-activated, gamma 3a non-catalytic subunit |
| chr13_-_29420885 | 1.03 |
ENSDART00000024225
|
chata
|
choline O-acetyltransferase a |
| chr17_-_29224908 | 1.02 |
ENSDART00000156288
|
si:dkey-28g23.6
|
si:dkey-28g23.6 |
| chr20_+_11731039 | 1.02 |
ENSDART00000152215
ENSDART00000152585 |
si:ch211-155o21.3
|
si:ch211-155o21.3 |
| chr2_+_33368414 | 1.02 |
ENSDART00000077462
|
slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr16_-_21140097 | 1.02 |
ENSDART00000145837
ENSDART00000146500 |
si:dkey-271j15.3
|
si:dkey-271j15.3 |
| chr16_-_45178430 | 1.01 |
ENSDART00000165186
|
si:dkey-33i11.9
|
si:dkey-33i11.9 |
| chr24_-_40860603 | 1.01 |
ENSDART00000188032
|
CU633479.7
|
|
| chr14_-_4145594 | 1.01 |
ENSDART00000077348
|
casp3b
|
caspase 3, apoptosis-related cysteine peptidase b |
| chr15_+_9327252 | 1.01 |
ENSDART00000144381
|
sgcg
|
sarcoglycan, gamma |
| chr24_-_7697274 | 1.00 |
ENSDART00000186077
|
syt5b
|
synaptotagmin Vb |
| chr15_-_46779934 | 0.99 |
ENSDART00000085136
|
clcn2c
|
chloride channel 2c |
| chr7_+_19495379 | 0.97 |
ENSDART00000180514
|
si:ch211-212k18.8
|
si:ch211-212k18.8 |
| chr3_-_19368435 | 0.96 |
ENSDART00000132987
|
s1pr5a
|
sphingosine-1-phosphate receptor 5a |
| chr22_+_38173960 | 0.96 |
ENSDART00000010537
|
cp
|
ceruloplasmin |
| chr15_-_37850969 | 0.96 |
ENSDART00000031418
|
hsc70
|
heat shock cognate 70 |
| chr22_-_24285432 | 0.95 |
ENSDART00000164083
|
si:ch211-117l17.4
|
si:ch211-117l17.4 |
| chr1_-_30723241 | 0.95 |
ENSDART00000152175
ENSDART00000152150 |
MZT1
|
si:dkey-15d12.2 |
| chr20_-_9462433 | 0.93 |
ENSDART00000152674
ENSDART00000040557 |
zgc:101840
|
zgc:101840 |
| chr7_-_71829649 | 0.91 |
ENSDART00000160449
|
cacnb2a
|
calcium channel, voltage-dependent, beta 2a |
| chr6_+_25257728 | 0.90 |
ENSDART00000162581
|
kyat3
|
kynurenine aminotransferase 3 |
| chr7_+_19495905 | 0.89 |
ENSDART00000125584
ENSDART00000173774 |
si:ch211-212k18.8
|
si:ch211-212k18.8 |
| chr1_-_5455498 | 0.89 |
ENSDART00000040368
ENSDART00000114035 |
mnx2b
|
motor neuron and pancreas homeobox 2b |
| chr20_+_52389858 | 0.89 |
ENSDART00000185863
ENSDART00000166651 |
arhgap39
|
Rho GTPase activating protein 39 |
| chr8_+_26141680 | 0.88 |
ENSDART00000078334
|
celsr3
|
cadherin, EGF LAG seven-pass G-type receptor 3 |
| chr6_+_7444899 | 0.88 |
ENSDART00000053775
|
arf3b
|
ADP-ribosylation factor 3b |
| chr11_+_40812590 | 0.88 |
ENSDART00000186690
|
errfi1a
|
ERBB receptor feedback inhibitor 1a |
| chr1_-_54063520 | 0.88 |
ENSDART00000171722
|
smdt1b
|
single-pass membrane protein with aspartate-rich tail 1b |
| chr1_-_42289704 | 0.87 |
ENSDART00000150124
|
si:ch211-71k14.1
|
si:ch211-71k14.1 |
| chr7_+_59020972 | 0.87 |
ENSDART00000157873
|
hacd1
|
3-hydroxyacyl-CoA dehydratase 1 |
| chr20_-_5291012 | 0.87 |
ENSDART00000122892
|
cyp46a1.3
|
cytochrome P450, family 46, subfamily A, polypeptide 1, tandem duplicate 3 |
| chr4_+_12111154 | 0.87 |
ENSDART00000036779
|
tmem178b
|
transmembrane protein 178B |
| chr17_+_38476300 | 0.85 |
ENSDART00000123298
|
stard9
|
StAR-related lipid transfer (START) domain containing 9 |
| chr18_+_19972853 | 0.84 |
ENSDART00000180071
|
skor1b
|
SKI family transcriptional corepressor 1b |
| chr1_-_22757145 | 0.83 |
ENSDART00000134719
|
prom1b
|
prominin 1 b |
| chr21_+_28478663 | 0.83 |
ENSDART00000077887
ENSDART00000134150 |
slc22a6l
|
solute carrier family 22 (organic anion transporter), member 6, like |
| chr20_-_22476255 | 0.83 |
ENSDART00000103510
|
pdgfra
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr8_+_31821396 | 0.81 |
ENSDART00000077053
|
plcxd3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr13_+_29771463 | 0.79 |
ENSDART00000134424
ENSDART00000138332 ENSDART00000134330 ENSDART00000160944 ENSDART00000076992 ENSDART00000160921 |
pax2a
|
paired box 2a |
| chr13_+_33282095 | 0.78 |
ENSDART00000135200
|
ccdc28b
|
coiled-coil domain containing 28B |
| chr3_-_36839115 | 0.78 |
ENSDART00000154553
|
ramp2
|
receptor (G protein-coupled) activity modifying protein 2 |
| chr11_+_41242644 | 0.77 |
ENSDART00000172008
|
pax7a
|
paired box 7a |
| chr8_-_13574764 | 0.76 |
ENSDART00000076561
|
B3GNT3 (1 of many)
|
si:ch211-126g16.10 |
| chr7_-_25895189 | 0.76 |
ENSDART00000173599
ENSDART00000079235 ENSDART00000173786 ENSDART00000173602 ENSDART00000079245 ENSDART00000187568 ENSDART00000173505 |
cd99l2
|
CD99 molecule-like 2 |
| chr24_-_6078222 | 0.76 |
ENSDART00000146830
|
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr9_-_51436377 | 0.75 |
ENSDART00000006612
|
tbr1b
|
T-box, brain, 1b |
| chr9_+_37152564 | 0.75 |
ENSDART00000189497
|
gli2a
|
GLI family zinc finger 2a |
| chr8_+_43340995 | 0.74 |
ENSDART00000038566
|
rflna
|
refilin A |
| chr14_+_4807207 | 0.74 |
ENSDART00000167145
|
ap1ar
|
adaptor-related protein complex 1 associated regulatory protein |
| chr21_-_5799122 | 0.74 |
ENSDART00000129351
ENSDART00000151202 |
ccni
|
cyclin I |
| chr25_-_6011034 | 0.73 |
ENSDART00000075197
ENSDART00000136054 |
snx22
|
sorting nexin 22 |
| chr14_-_17306261 | 0.73 |
ENSDART00000191747
|
jakmip1
|
janus kinase and microtubule interacting protein 1 |
| chr18_-_38088099 | 0.73 |
ENSDART00000146120
|
luzp2
|
leucine zipper protein 2 |
| chr16_-_28878080 | 0.72 |
ENSDART00000149501
|
si:dkey-239n17.4
|
si:dkey-239n17.4 |
| chr23_-_16485190 | 0.72 |
ENSDART00000155038
|
si:dkeyp-100a5.4
|
si:dkeyp-100a5.4 |
| chr4_+_19700308 | 0.72 |
ENSDART00000027919
|
pax4
|
paired box 4 |
| chr20_+_7584211 | 0.72 |
ENSDART00000132481
ENSDART00000127975 ENSDART00000144551 |
bloc1s2
|
biogenesis of lysosomal organelles complex-1, subunit 2 |
| chr16_+_28728347 | 0.72 |
ENSDART00000149240
|
si:dkey-24i24.3
|
si:dkey-24i24.3 |
| chr17_-_42213822 | 0.72 |
ENSDART00000187904
ENSDART00000180029 |
nkx2.2a
|
NK2 homeobox 2a |
| chr5_-_54672763 | 0.72 |
ENSDART00000159009
|
spag8
|
sperm associated antigen 8 |
| chr19_+_43780970 | 0.71 |
ENSDART00000063870
|
rpl11
|
ribosomal protein L11 |
| chr7_+_17229980 | 0.71 |
ENSDART00000184910
|
slc6a5
|
solute carrier family 6 (neurotransmitter transporter), member 5 |
| chr15_+_36309070 | 0.71 |
ENSDART00000157034
|
gmnc
|
geminin coiled-coil domain containing |
| chr1_+_51721851 | 0.70 |
ENSDART00000040397
|
prdx2
|
peroxiredoxin 2 |
| chr9_-_31278048 | 0.70 |
ENSDART00000022204
|
zic5
|
zic family member 5 (odd-paired homolog, Drosophila) |
| chr21_-_17482465 | 0.69 |
ENSDART00000004548
|
barhl1b
|
BarH-like homeobox 1b |
| chr8_-_34051548 | 0.69 |
ENSDART00000105204
|
pbx3b
|
pre-B-cell leukemia homeobox 3b |
| chr4_-_4387012 | 0.69 |
ENSDART00000191836
|
CU468826.3
|
Danio rerio U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit-related protein 1-like (LOC100331497), mRNA. |
| chr1_+_23784905 | 0.68 |
ENSDART00000171951
ENSDART00000188521 ENSDART00000183029 ENSDART00000187183 |
slit2
|
slit homolog 2 (Drosophila) |
| chr15_+_27384798 | 0.68 |
ENSDART00000164887
|
tbx4
|
T-box 4 |
| chr7_-_68373495 | 0.67 |
ENSDART00000167440
|
zfhx3
|
zinc finger homeobox 3 |
| chr15_+_9861973 | 0.67 |
ENSDART00000170945
|
si:dkey-13m3.2
|
si:dkey-13m3.2 |
| chr17_+_16564921 | 0.67 |
ENSDART00000151904
|
foxn3
|
forkhead box N3 |
| chr6_-_28980756 | 0.66 |
ENSDART00000014661
|
glmnb
|
glomulin, FKBP associated protein b |
| chr8_-_31062811 | 0.66 |
ENSDART00000142528
|
slc20a1a
|
solute carrier family 20, member 1a |
| chr14_-_7137808 | 0.66 |
ENSDART00000054803
|
trpt1
|
tRNA phosphotransferase 1 |
| chr19_-_11106315 | 0.65 |
ENSDART00000059102
|
arhgef1a
|
Rho guanine nucleotide exchange factor (GEF) 1a |
| chr13_+_5570952 | 0.65 |
ENSDART00000134506
|
si:dkey-196j8.2
|
si:dkey-196j8.2 |
| chr14_-_8080416 | 0.65 |
ENSDART00000045109
|
zgc:92242
|
zgc:92242 |
| chr20_-_2949028 | 0.64 |
ENSDART00000104667
ENSDART00000193151 ENSDART00000131946 |
cdk19
|
cyclin-dependent kinase 19 |
| chr25_-_13839743 | 0.64 |
ENSDART00000158780
|
mapk8ip1a
|
mitogen-activated protein kinase 8 interacting protein 1a |
| chr6_-_8311044 | 0.64 |
ENSDART00000129674
|
slc44a2
|
solute carrier family 44 (choline transporter), member 2 |
| chr17_+_11675362 | 0.63 |
ENSDART00000157911
|
kif26ba
|
kinesin family member 26Ba |
| chr13_-_29421331 | 0.62 |
ENSDART00000150228
|
chata
|
choline O-acetyltransferase a |
| chr18_-_48547564 | 0.62 |
ENSDART00000138607
|
kcnj1a.1
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 1 |
| chr16_+_28932038 | 0.62 |
ENSDART00000149480
|
npr1b
|
natriuretic peptide receptor 1b |
| chr23_-_20051369 | 0.61 |
ENSDART00000049836
|
bgnb
|
biglycan b |
| chr3_-_32337653 | 0.60 |
ENSDART00000156918
ENSDART00000156551 |
si:dkey-16p21.8
|
si:dkey-16p21.8 |
| chr11_-_45141309 | 0.60 |
ENSDART00000181736
|
cant1b
|
calcium activated nucleotidase 1b |
| chr8_+_8459192 | 0.60 |
ENSDART00000140942
ENSDART00000014939 |
comta
|
catechol-O-methyltransferase a |
| chr13_-_2010191 | 0.60 |
ENSDART00000161021
ENSDART00000124134 |
gfral
|
GDNF family receptor alpha like |
| chr25_+_7671640 | 0.59 |
ENSDART00000145367
|
kcnj11l
|
potassium inwardly-rectifying channel, subfamily J, member 11, like |
| chr3_-_12970418 | 0.59 |
ENSDART00000158747
|
pdgfab
|
platelet-derived growth factor alpha polypeptide b |
| chr18_-_17030870 | 0.58 |
ENSDART00000079817
|
has3
|
hyaluronan synthase 3 |
| chr15_-_8309207 | 0.58 |
ENSDART00000143880
ENSDART00000061351 |
tnfrsf19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr14_-_15956921 | 0.58 |
ENSDART00000190500
|
flt4
|
fms-related tyrosine kinase 4 |
| chr14_-_858985 | 0.58 |
ENSDART00000148687
ENSDART00000149375 |
slc34a1a
|
solute carrier family 34 (type II sodium/phosphate cotransporter), member 1a |
| chr4_-_9909371 | 0.58 |
ENSDART00000102656
|
si:dkey-22l11.6
|
si:dkey-22l11.6 |
| chr4_+_16715267 | 0.57 |
ENSDART00000143849
|
pkp2
|
plakophilin 2 |
| chr15_-_20939579 | 0.57 |
ENSDART00000152371
|
usp2a
|
ubiquitin specific peptidase 2a |
| chr10_+_22381802 | 0.57 |
ENSDART00000112484
|
nlgn2b
|
neuroligin 2b |
| chr2_-_59327299 | 0.56 |
ENSDART00000133734
|
ftr36
|
finTRIM family, member 36 |
| chr19_-_28130658 | 0.56 |
ENSDART00000079114
|
irx1b
|
iroquois homeobox 1b |
| chr16_-_26296477 | 0.55 |
ENSDART00000157553
|
erfl1
|
Ets2 repressor factor like 1 |
| chr20_-_28642061 | 0.54 |
ENSDART00000135513
|
rgs6
|
regulator of G protein signaling 6 |
| chr7_+_66822229 | 0.54 |
ENSDART00000112109
|
lyve1a
|
lymphatic vessel endothelial hyaluronic receptor 1a |
| chr22_-_26865361 | 0.54 |
ENSDART00000182504
|
hmox2a
|
heme oxygenase 2a |
| chr1_+_44127292 | 0.54 |
ENSDART00000160542
|
cabp2a
|
calcium binding protein 2a |
| chr23_-_18913032 | 0.54 |
ENSDART00000136678
|
si:ch211-209j10.6
|
si:ch211-209j10.6 |
| chr3_-_47876427 | 0.53 |
ENSDART00000180844
ENSDART00000124480 |
adgrl1a
|
adhesion G protein-coupled receptor L1a |
| chr16_+_46401576 | 0.53 |
ENSDART00000130264
|
rpz
|
rapunzel |
| chr14_-_451555 | 0.53 |
ENSDART00000190906
|
FAT4
|
FAT atypical cadherin 4 |
| chr10_-_27049170 | 0.52 |
ENSDART00000143451
|
cnih2
|
cornichon family AMPA receptor auxiliary protein 2 |
| chr24_+_39108243 | 0.52 |
ENSDART00000156353
|
mss51
|
MSS51 mitochondrial translational activator |
| chr20_+_25225112 | 0.52 |
ENSDART00000153088
ENSDART00000127291 ENSDART00000130494 |
moxd1
|
monooxygenase, DBH-like 1 |
| chr3_-_29941357 | 0.51 |
ENSDART00000147732
ENSDART00000137973 ENSDART00000103523 |
grna
|
granulin a |
| chr1_-_45177373 | 0.51 |
ENSDART00000143142
ENSDART00000034549 |
zgc:111983
|
zgc:111983 |
| chr7_+_20475788 | 0.51 |
ENSDART00000171155
|
si:dkey-19b23.13
|
si:dkey-19b23.13 |
| chr15_-_18162647 | 0.50 |
ENSDART00000012064
|
pih1d2
|
PIH1 domain containing 2 |
| chr25_+_13620555 | 0.49 |
ENSDART00000163642
|
si:ch211-172l8.4
|
si:ch211-172l8.4 |
| chr8_-_12867434 | 0.49 |
ENSDART00000081657
|
slc2a6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr18_-_17485419 | 0.49 |
ENSDART00000018764
|
foxl1
|
forkhead box L1 |
| chr8_+_2487250 | 0.49 |
ENSDART00000081325
|
dynll1
|
dynein, light chain, LC8-type 1 |
| chr8_+_18830759 | 0.49 |
ENSDART00000089079
|
mpnd
|
MPN domain containing |
Network of associatons between targets according to the STRING database.
First level regulatory network of lhx3+lhx4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.0 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) negative regulation of striated muscle contraction(GO:0045988) relaxation of skeletal muscle(GO:0090076) |
| 0.6 | 1.9 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.6 | 1.9 | GO:0021530 | spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.6 | 1.7 | GO:0008292 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.5 | 2.0 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.4 | 2.9 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.3 | 2.7 | GO:0030279 | negative regulation of ossification(GO:0030279) |
| 0.3 | 1.6 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.3 | 1.0 | GO:0060092 | regulation of synaptic transmission, glycinergic(GO:0060092) |
| 0.3 | 1.3 | GO:0046462 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.2 | 1.2 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.2 | 0.7 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
| 0.2 | 0.7 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.2 | 0.6 | GO:0015871 | choline transport(GO:0015871) |
| 0.2 | 0.6 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.2 | 0.7 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.2 | 1.1 | GO:0042424 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.2 | 0.7 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) |
| 0.2 | 1.9 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.2 | 1.9 | GO:0021703 | locus ceruleus development(GO:0021703) |
| 0.2 | 0.8 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.2 | 1.3 | GO:0043584 | nose development(GO:0043584) |
| 0.2 | 1.5 | GO:0040014 | regulation of multicellular organism growth(GO:0040014) |
| 0.2 | 1.1 | GO:0042983 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.2 | 0.8 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 0.2 | 6.3 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 0.4 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.1 | 0.4 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.1 | 1.8 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.1 | 0.6 | GO:0014045 | establishment of endothelial blood-brain barrier(GO:0014045) |
| 0.1 | 0.7 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.1 | 0.7 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.1 | 0.4 | GO:0060898 | eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.1 | 0.9 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.1 | 0.5 | GO:0060118 | vestibular receptor cell differentiation(GO:0060114) vestibular receptor cell development(GO:0060118) |
| 0.1 | 1.7 | GO:0032309 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 | 0.7 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.1 | 0.7 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.6 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.1 | 1.0 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.1 | 0.7 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.1 | 0.6 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.1 | 4.5 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.1 | 0.3 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.1 | 2.9 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.1 | 0.5 | GO:0097272 | ammonia homeostasis(GO:0097272) |
| 0.1 | 0.7 | GO:0043092 | amino acid import(GO:0043090) L-amino acid import(GO:0043092) |
| 0.1 | 0.6 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.1 | 0.6 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.1 | 0.3 | GO:0015919 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.1 | 0.7 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.1 | 0.9 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 | 1.4 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.1 | 0.6 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.1 | 1.3 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 | 0.5 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 0.7 | GO:0098869 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.1 | 2.6 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 0.9 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 1.4 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 1.2 | GO:0000302 | response to reactive oxygen species(GO:0000302) |
| 0.1 | 0.7 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 0.5 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.1 | 0.4 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 1.9 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.1 | 1.0 | GO:0042026 | protein refolding(GO:0042026) |
| 0.1 | 2.3 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.1 | 0.6 | GO:0097105 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.1 | 0.6 | GO:0071678 | olfactory bulb axon guidance(GO:0071678) |
| 0.1 | 0.4 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.1 | 0.2 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.1 | 1.5 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.1 | 0.9 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 0.2 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.0 | 0.3 | GO:0090497 | mesenchymal cell migration(GO:0090497) |
| 0.0 | 0.9 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.2 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.5 | GO:0060122 | inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 | 1.1 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.7 | GO:1902287 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 1.9 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.9 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.0 | 0.5 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 2.2 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.4 | GO:0000272 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.1 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) |
| 0.0 | 0.1 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.0 | 0.7 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 0.7 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 3.8 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.0 | 0.9 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 1.3 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.0 | 0.1 | GO:0046083 | adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.0 | 0.8 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.1 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.0 | 0.4 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 1.5 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.4 | GO:1903672 | positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.0 | 1.2 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 0.0 | 0.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.0 | 1.2 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.2 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.5 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.0 | 0.8 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 1.1 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.7 | GO:0035138 | pectoral fin morphogenesis(GO:0035138) |
| 0.0 | 0.7 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 1.6 | GO:0061515 | myeloid cell development(GO:0061515) |
| 0.0 | 0.6 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.3 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.1 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.0 | 0.2 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.2 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.4 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.4 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.4 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.0 | 0.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.1 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.0 | 0.1 | GO:0034982 | mitochondrial protein processing(GO:0034982) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.0 | GO:0031673 | H zone(GO:0031673) |
| 0.3 | 1.9 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.2 | 2.6 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.2 | 0.6 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.2 | 2.9 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.2 | 6.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 1.0 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.5 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 0.8 | GO:0071914 | prominosome(GO:0071914) |
| 0.1 | 1.0 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 1.1 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 1.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 2.9 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 0.9 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 0.7 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.3 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.6 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 1.5 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.4 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.4 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.7 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.6 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.5 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.3 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.4 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 1.4 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 2.2 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.1 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.7 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.6 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 1.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.9 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.4 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 2.1 | GO:0005635 | nuclear envelope(GO:0005635) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0001605 | adrenomedullin receptor activity(GO:0001605) calcitonin gene-related peptide receptor activity(GO:0001635) |
| 0.4 | 5.8 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.4 | 3.1 | GO:0036122 | BMP binding(GO:0036122) |
| 0.2 | 0.7 | GO:0055105 | ubiquitin-protein transferase inhibitor activity(GO:0055105) |
| 0.2 | 0.9 | GO:0018812 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.2 | 0.8 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 0.6 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.2 | 0.7 | GO:0031005 | filamin binding(GO:0031005) |
| 0.2 | 1.2 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.2 | 3.0 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.2 | 0.5 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.2 | 0.6 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.2 | 0.5 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) |
| 0.1 | 0.9 | GO:0033781 | cholesterol 24-hydroxylase activity(GO:0033781) |
| 0.1 | 1.6 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 1.5 | GO:0031995 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.4 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.1 | 1.7 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 1.7 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 0.6 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.1 | 1.0 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 0.7 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 0.7 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 0.4 | GO:0016521 | pituitary adenylate cyclase activating polypeptide activity(GO:0016521) |
| 0.1 | 0.7 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 1.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.6 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.1 | 0.6 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 0.4 | GO:0004135 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.1 | 2.6 | GO:0016208 | AMP binding(GO:0016208) |
| 0.1 | 1.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 0.4 | GO:0098634 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 0.2 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.1 | 0.4 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 0.5 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 0.6 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 1.0 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 1.0 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.6 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.9 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 0.2 | GO:0034618 | arginine binding(GO:0034618) |
| 0.1 | 1.0 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 1.8 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.3 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 2.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.6 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 1.1 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 1.0 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 0.7 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.7 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.7 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.8 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 1.8 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 1.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 1.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.7 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 1.0 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.7 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.0 | 0.7 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.3 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 2.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.6 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.0 | 0.9 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.2 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 2.5 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.4 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 0.5 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.7 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.9 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.2 | GO:0005351 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 2.0 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.1 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.0 | 0.1 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.0 | 1.5 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.4 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.4 | GO:0098631 | protein binding involved in cell adhesion(GO:0098631) protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.1 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.8 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.5 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 26.8 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.0 | 0.3 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 0.6 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.3 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.7 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 2.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.0 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.8 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.4 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.7 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 2.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.0 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 2.4 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 0.7 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 0.6 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 1.9 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 0.7 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 0.8 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.6 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.2 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 1.0 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 1.2 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.7 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.3 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.4 | REACTOME TRANSPORT OF GLUCOSE AND OTHER SUGARS BILE SALTS AND ORGANIC ACIDS METAL IONS AND AMINE COMPOUNDS | Genes involved in Transport of glucose and other sugars, bile salts and organic acids, metal ions and amine compounds |
| 0.0 | 0.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.1 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.1 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |