Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
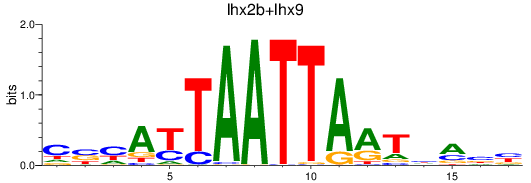
Results for lhx2b+lhx9
Z-value: 1.07
Transcription factors associated with lhx2b+lhx9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
lhx2b
|
ENSDARG00000031222 | LIM homeobox 2b |
|
lhx9
|
ENSDARG00000056979 | LIM homeobox 9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| lhx9 | dr11_v1_chr22_-_23253252_23253252 | -0.96 | 5.6e-05 | Click! |
| lhx2b | dr11_v1_chr8_+_3085120_3085219 | -0.82 | 7.2e-03 | Click! |
Activity profile of lhx2b+lhx9 motif
Sorted Z-values of lhx2b+lhx9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_+_22320738 | 2.42 |
ENSDART00000073566
|
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr17_-_4245902 | 2.15 |
ENSDART00000151851
|
gdf3
|
growth differentiation factor 3 |
| chr21_+_25777425 | 2.12 |
ENSDART00000021620
|
cldnd
|
claudin d |
| chr10_-_34916208 | 2.08 |
ENSDART00000187371
|
ccna1
|
cyclin A1 |
| chr8_+_45334255 | 2.00 |
ENSDART00000126848
ENSDART00000134161 ENSDART00000142322 ENSDART00000145011 ENSDART00000183560 |
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr8_-_53044300 | 1.79 |
ENSDART00000191653
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr9_+_48761455 | 1.69 |
ENSDART00000139631
|
abcb11a
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11a |
| chr25_-_21031007 | 1.63 |
ENSDART00000138985
|
gnaia
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a |
| chr14_-_33481428 | 1.58 |
ENSDART00000147059
ENSDART00000140001 ENSDART00000124242 ENSDART00000164836 ENSDART00000190104 ENSDART00000186833 ENSDART00000180873 |
lamp2
|
lysosomal-associated membrane protein 2 |
| chr19_+_2631565 | 1.39 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr10_-_34002185 | 1.28 |
ENSDART00000046599
|
zar1l
|
zygote arrest 1-like |
| chr23_-_18024543 | 1.16 |
ENSDART00000139695
|
pm20d1.1
|
peptidase M20 domain containing 1, tandem duplicate 1 |
| chr16_-_42965192 | 1.14 |
ENSDART00000113714
|
mtx1a
|
metaxin 1a |
| chr1_-_55248496 | 1.14 |
ENSDART00000098615
|
nanos3
|
nanos homolog 3 |
| chr8_-_25771474 | 1.12 |
ENSDART00000193883
|
suv39h1b
|
suppressor of variegation 3-9 homolog 1b |
| chr24_-_26622423 | 1.12 |
ENSDART00000182044
|
tnikb
|
TRAF2 and NCK interacting kinase b |
| chr22_+_23359369 | 1.07 |
ENSDART00000170886
|
dennd1b
|
DENN/MADD domain containing 1B |
| chr1_+_35985813 | 1.06 |
ENSDART00000179634
ENSDART00000139636 ENSDART00000175902 |
zgc:152968
|
zgc:152968 |
| chr21_-_44081540 | 1.06 |
ENSDART00000130833
|
FO704810.1
|
|
| chr8_-_20230559 | 1.05 |
ENSDART00000193677
|
mllt1a
|
MLLT1, super elongation complex subunit a |
| chr2_-_9989919 | 1.02 |
ENSDART00000180213
ENSDART00000184369 |
imp3
|
IMP3, U3 small nucleolar ribonucleoprotein, homolog (yeast) |
| chr5_-_32396929 | 1.01 |
ENSDART00000023977
|
fbxw2
|
F-box and WD repeat domain containing 2 |
| chr5_+_37903790 | 0.99 |
ENSDART00000162470
|
tmprss4b
|
transmembrane protease, serine 4b |
| chr7_+_6941583 | 0.98 |
ENSDART00000160709
ENSDART00000157634 |
rbm14b
|
RNA binding motif protein 14b |
| chr12_-_18578218 | 0.93 |
ENSDART00000125803
|
zdhhc4
|
zinc finger, DHHC-type containing 4 |
| chr20_-_49889111 | 0.92 |
ENSDART00000058858
|
kif13bb
|
kinesin family member 13Bb |
| chr18_+_35128685 | 0.90 |
ENSDART00000151579
|
si:ch211-195m9.3
|
si:ch211-195m9.3 |
| chr23_+_4709607 | 0.89 |
ENSDART00000166503
ENSDART00000158752 ENSDART00000163860 ENSDART00000172739 |
raf1a
raf1a
|
Raf-1 proto-oncogene, serine/threonine kinase a Raf-1 proto-oncogene, serine/threonine kinase a |
| chr5_+_63302660 | 0.87 |
ENSDART00000142131
|
si:ch73-376l24.2
|
si:ch73-376l24.2 |
| chr17_+_24318753 | 0.86 |
ENSDART00000064083
|
otx1
|
orthodenticle homeobox 1 |
| chr23_-_19225709 | 0.86 |
ENSDART00000080099
|
oard1
|
O-acyl-ADP-ribose deacylase 1 |
| chr1_+_418869 | 0.85 |
ENSDART00000152173
|
tpp2
|
tripeptidyl peptidase 2 |
| chr21_+_21195487 | 0.83 |
ENSDART00000181746
ENSDART00000184832 |
rictorb
|
RPTOR independent companion of MTOR, complex 2b |
| chr5_+_29851433 | 0.82 |
ENSDART00000143434
|
ubash3ba
|
ubiquitin associated and SH3 domain containing Ba |
| chr12_-_18578432 | 0.82 |
ENSDART00000122858
|
zdhhc4
|
zinc finger, DHHC-type containing 4 |
| chr10_+_33393829 | 0.81 |
ENSDART00000163458
ENSDART00000115379 |
zgc:153345
|
zgc:153345 |
| chr13_+_35528607 | 0.81 |
ENSDART00000075414
ENSDART00000112947 |
wdr27
|
WD repeat domain 27 |
| chr9_-_43644261 | 0.79 |
ENSDART00000023684
|
cwc22
|
CWC22 spliceosome-associated protein homolog (S. cerevisiae) |
| chr2_-_17392799 | 0.79 |
ENSDART00000136470
ENSDART00000141188 |
st3gal3b
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3b |
| chr8_-_20230802 | 0.79 |
ENSDART00000063400
|
mllt1a
|
MLLT1, super elongation complex subunit a |
| chr18_+_14684115 | 0.79 |
ENSDART00000108469
|
spata2l
|
spermatogenesis associated 2-like |
| chr2_-_17393216 | 0.78 |
ENSDART00000123137
|
st3gal3b
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3b |
| chr5_-_11809710 | 0.78 |
ENSDART00000186998
ENSDART00000181363 ENSDART00000180681 |
nf2a
|
neurofibromin 2a (merlin) |
| chr7_+_57088920 | 0.78 |
ENSDART00000024076
|
scamp2l
|
secretory carrier membrane protein 2, like |
| chr17_-_51893123 | 0.76 |
ENSDART00000103350
ENSDART00000017329 |
numb
|
numb homolog (Drosophila) |
| chr6_-_8597735 | 0.76 |
ENSDART00000151294
|
fopnl
|
fgfr1op N-terminal like |
| chr22_-_20924564 | 0.76 |
ENSDART00000100642
ENSDART00000032770 |
ell
|
elongation factor RNA polymerase II |
| chr17_-_40956035 | 0.75 |
ENSDART00000124715
|
si:dkey-16j16.4
|
si:dkey-16j16.4 |
| chr23_-_33679579 | 0.74 |
ENSDART00000188674
|
tfcp2
|
transcription factor CP2 |
| chr6_-_15491579 | 0.74 |
ENSDART00000156439
|
st6gal2b
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 2b |
| chr23_+_1029450 | 0.74 |
ENSDART00000189196
|
si:zfos-905g2.1
|
si:zfos-905g2.1 |
| chr21_-_13123176 | 0.74 |
ENSDART00000144866
ENSDART00000024616 |
fam219aa
|
family with sequence similarity 219, member Aa |
| chr17_-_27048537 | 0.72 |
ENSDART00000050018
ENSDART00000193861 |
cnksr1
|
connector enhancer of kinase suppressor of Ras 1 |
| chr6_-_15492030 | 0.72 |
ENSDART00000156141
ENSDART00000183992 |
st6gal2b
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 2b |
| chr5_-_66397688 | 0.70 |
ENSDART00000161483
|
hip1rb
|
huntingtin interacting protein 1 related b |
| chr10_-_13343831 | 0.69 |
ENSDART00000135941
|
il11ra
|
interleukin 11 receptor, alpha |
| chr16_-_31622777 | 0.69 |
ENSDART00000137311
ENSDART00000002930 |
phf20l1
|
PHD finger protein 20 like 1 |
| chr21_-_2814709 | 0.68 |
ENSDART00000097664
|
SEMA4D
|
semaphorin 4D |
| chr19_-_7441686 | 0.67 |
ENSDART00000168194
|
gabpb2a
|
GA binding protein transcription factor, beta subunit 2a |
| chr2_+_10878406 | 0.66 |
ENSDART00000091497
|
tceanc2
|
transcription elongation factor A (SII) N-terminal and central domain containing 2 |
| chr5_+_872299 | 0.66 |
ENSDART00000130042
|
fubp3
|
far upstream element (FUSE) binding protein 3 |
| chr22_-_20812822 | 0.66 |
ENSDART00000193778
|
dot1l
|
DOT1-like histone H3K79 methyltransferase |
| chr23_-_25779995 | 0.65 |
ENSDART00000110670
|
si:dkey-21c19.3
|
si:dkey-21c19.3 |
| chr21_+_43172506 | 0.65 |
ENSDART00000121725
|
zcchc10
|
zinc finger, CCHC domain containing 10 |
| chr21_+_13387965 | 0.64 |
ENSDART00000134347
|
zgc:113162
|
zgc:113162 |
| chr5_-_57723929 | 0.64 |
ENSDART00000144237
|
gig2p
|
grass carp reovirus (GCRV)-induced gene 2p |
| chr24_+_17260329 | 0.64 |
ENSDART00000129554
|
bmi1a
|
bmi1 polycomb ring finger oncogene 1a |
| chr20_-_51831816 | 0.64 |
ENSDART00000060505
|
mia3
|
melanoma inhibitory activity family, member 3 |
| chr5_+_9224051 | 0.63 |
ENSDART00000139265
|
si:ch211-12e13.12
|
si:ch211-12e13.12 |
| chr24_+_17260001 | 0.63 |
ENSDART00000066765
|
bmi1a
|
bmi1 polycomb ring finger oncogene 1a |
| chr19_+_1688727 | 0.63 |
ENSDART00000115136
ENSDART00000166744 |
dennd3a
|
DENN/MADD domain containing 3a |
| chr5_-_48664522 | 0.63 |
ENSDART00000083229
|
mblac2
|
metallo-beta-lactamase domain containing 2 |
| chr2_+_9990491 | 0.61 |
ENSDART00000011906
|
slc35a3b
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3b |
| chr11_+_17984354 | 0.61 |
ENSDART00000179986
|
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr14_-_14659023 | 0.61 |
ENSDART00000170355
ENSDART00000159888 ENSDART00000172241 |
nsdhl
|
NAD(P) dependent steroid dehydrogenase-like |
| chr19_-_27391179 | 0.60 |
ENSDART00000181108
|
mgat1b
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase b |
| chr6_+_12527725 | 0.59 |
ENSDART00000149328
|
stk24b
|
serine/threonine kinase 24b (STE20 homolog, yeast) |
| chr15_-_30505607 | 0.59 |
ENSDART00000155212
|
msi2b
|
musashi RNA-binding protein 2b |
| chr10_-_31015535 | 0.58 |
ENSDART00000146116
|
panx3
|
pannexin 3 |
| chr17_-_25831569 | 0.58 |
ENSDART00000148743
|
hhat
|
hedgehog acyltransferase |
| chr5_-_18961694 | 0.58 |
ENSDART00000142531
ENSDART00000090521 |
ankle2
|
ankyrin repeat and LEM domain containing 2 |
| chr3_+_53352018 | 0.58 |
ENSDART00000082715
|
camsap3
|
calmodulin regulated spectrin-associated protein family, member 3 |
| chr19_+_15485287 | 0.57 |
ENSDART00000182797
|
pdik1l
|
PDLIM1 interacting kinase 1 like |
| chr9_+_54039006 | 0.57 |
ENSDART00000112441
|
tlr7
|
toll-like receptor 7 |
| chr5_+_13394543 | 0.57 |
ENSDART00000051669
ENSDART00000135921 |
tctn2
|
tectonic family member 2 |
| chr12_+_47698356 | 0.57 |
ENSDART00000112010
|
lzts2b
|
leucine zipper, putative tumor suppressor 2b |
| chr15_+_34934568 | 0.57 |
ENSDART00000165210
|
zgc:66024
|
zgc:66024 |
| chr15_-_38129845 | 0.56 |
ENSDART00000057095
|
si:dkey-24p1.1
|
si:dkey-24p1.1 |
| chr23_-_2901167 | 0.56 |
ENSDART00000165955
ENSDART00000190616 |
zhx3
|
zinc fingers and homeoboxes 3 |
| chr9_+_24065855 | 0.56 |
ENSDART00000161468
ENSDART00000171577 ENSDART00000172743 ENSDART00000159324 ENSDART00000079689 ENSDART00000023196 ENSDART00000101577 |
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr1_+_47499888 | 0.56 |
ENSDART00000027624
|
stn1
|
STN1, CST complex subunit |
| chr9_+_28150275 | 0.56 |
ENSDART00000192129
|
plekhm3
|
pleckstrin homology domain containing, family M, member 3 |
| chr2_+_105748 | 0.56 |
ENSDART00000169601
|
CABZ01098670.1
|
|
| chr20_+_5985329 | 0.55 |
ENSDART00000165489
|
cep128
|
centrosomal protein 128 |
| chr8_-_51930826 | 0.55 |
ENSDART00000109785
|
cabin1
|
calcineurin binding protein 1 |
| chr7_+_23515966 | 0.55 |
ENSDART00000186893
ENSDART00000186189 |
zgc:109889
|
zgc:109889 |
| chr9_+_7732714 | 0.55 |
ENSDART00000145853
|
si:ch1073-349o24.2
|
si:ch1073-349o24.2 |
| chr12_-_4243268 | 0.55 |
ENSDART00000131275
|
zgc:92313
|
zgc:92313 |
| chr20_-_45060241 | 0.54 |
ENSDART00000185227
|
klhl29
|
kelch-like family member 29 |
| chr4_+_6869847 | 0.54 |
ENSDART00000036646
|
dock4b
|
dedicator of cytokinesis 4b |
| chr17_-_16422654 | 0.54 |
ENSDART00000150149
|
tdp1
|
tyrosyl-DNA phosphodiesterase 1 |
| chr23_-_36303216 | 0.54 |
ENSDART00000188720
|
cbx5
|
chromobox homolog 5 (HP1 alpha homolog, Drosophila) |
| chr10_+_17235370 | 0.53 |
ENSDART00000038780
|
sppl3
|
signal peptide peptidase 3 |
| chr12_-_18577983 | 0.53 |
ENSDART00000193262
|
zdhhc4
|
zinc finger, DHHC-type containing 4 |
| chr8_+_36560019 | 0.53 |
ENSDART00000136418
ENSDART00000061378 ENSDART00000185237 |
sf3a1
|
splicing factor 3a, subunit 1 |
| chr20_-_31308805 | 0.52 |
ENSDART00000147045
|
hpcal1
|
hippocalcin-like 1 |
| chr14_-_470505 | 0.52 |
ENSDART00000067147
|
ANKRD50
|
ankyrin repeat domain 50 |
| chr6_+_40922572 | 0.52 |
ENSDART00000133599
ENSDART00000002728 ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr3_-_59297532 | 0.51 |
ENSDART00000187991
|
CABZ01053748.1
|
|
| chr21_-_25801956 | 0.50 |
ENSDART00000101219
|
mettl27
|
methyltransferase like 27 |
| chr23_+_20705849 | 0.49 |
ENSDART00000079538
|
ccdc30
|
coiled-coil domain containing 30 |
| chr19_-_7441948 | 0.47 |
ENSDART00000003544
|
gabpb2a
|
GA binding protein transcription factor, beta subunit 2a |
| chr23_+_384850 | 0.47 |
ENSDART00000114000
|
zgc:101663
|
zgc:101663 |
| chr5_+_1624359 | 0.47 |
ENSDART00000165431
|
PPP1CC
|
protein phosphatase 1 catalytic subunit gamma |
| chr6_-_1432200 | 0.47 |
ENSDART00000182901
|
LO018148.1
|
|
| chr20_-_43663494 | 0.46 |
ENSDART00000144564
|
BX470188.1
|
|
| chr20_+_28364742 | 0.45 |
ENSDART00000103355
|
rhov
|
ras homolog family member V |
| chr9_-_21488976 | 0.45 |
ENSDART00000080404
|
mphosph8
|
M-phase phosphoprotein 8 |
| chr19_-_30800004 | 0.45 |
ENSDART00000128560
ENSDART00000045504 ENSDART00000125893 |
trit1
|
tRNA isopentenyltransferase 1 |
| chr10_+_44373349 | 0.45 |
ENSDART00000172191
|
snrnp35
|
small nuclear ribonucleoprotein 35 (U11/U12) |
| chr13_-_31008275 | 0.45 |
ENSDART00000139394
|
wdfy4
|
WDFY family member 4 |
| chr7_-_51727760 | 0.44 |
ENSDART00000174180
|
hdac8
|
histone deacetylase 8 |
| chr11_+_17984167 | 0.44 |
ENSDART00000020283
ENSDART00000188329 |
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr20_+_98179 | 0.44 |
ENSDART00000022725
|
si:ch1073-155h21.1
|
si:ch1073-155h21.1 |
| chr15_+_21262917 | 0.44 |
ENSDART00000101000
|
gkup
|
glucuronokinase with putative uridyl pyrophosphorylase |
| chr1_-_45616470 | 0.44 |
ENSDART00000150165
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr22_+_508290 | 0.43 |
ENSDART00000135403
|
nuak2
|
NUAK family, SNF1-like kinase, 2 |
| chr11_+_45436703 | 0.43 |
ENSDART00000168295
ENSDART00000173293 |
sos1
|
son of sevenless homolog 1 (Drosophila) |
| chr13_-_38730267 | 0.43 |
ENSDART00000157524
|
lmbrd1
|
LMBR1 domain containing 1 |
| chr21_+_6394929 | 0.43 |
ENSDART00000138600
|
si:ch211-225g23.1
|
si:ch211-225g23.1 |
| chr13_+_18321140 | 0.42 |
ENSDART00000180947
|
eif4e1c
|
eukaryotic translation initiation factor 4E family member 1c |
| chr18_-_16885362 | 0.42 |
ENSDART00000132778
|
swap70b
|
switching B cell complex subunit SWAP70b |
| chr12_+_47081783 | 0.42 |
ENSDART00000158568
|
mtr
|
5-methyltetrahydrofolate-homocysteine methyltransferase |
| chr15_-_43284021 | 0.42 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr8_+_7801060 | 0.42 |
ENSDART00000161618
|
tfe3a
|
transcription factor binding to IGHM enhancer 3a |
| chr10_-_2788668 | 0.42 |
ENSDART00000131749
ENSDART00000124356 ENSDART00000085031 |
ash2l
|
ash2 (absent, small, or homeotic)-like (Drosophila) |
| chr10_+_40324395 | 0.41 |
ENSDART00000147205
|
gltpb
|
glycolipid transfer protein b |
| chr9_+_48123224 | 0.41 |
ENSDART00000141610
|
klhl23
|
kelch-like family member 23 |
| chr13_+_30572172 | 0.41 |
ENSDART00000010052
ENSDART00000144417 |
ppifa
|
peptidylprolyl isomerase Fa |
| chr18_+_17537344 | 0.41 |
ENSDART00000025782
|
nup93
|
nucleoporin 93 |
| chr11_-_6068375 | 0.41 |
ENSDART00000167672
ENSDART00000122262 |
babam1
|
BRISC and BRCA1 A complex member 1 |
| chr11_+_42726712 | 0.40 |
ENSDART00000028955
|
tdrd3
|
tudor domain containing 3 |
| chr25_-_25058508 | 0.40 |
ENSDART00000087570
ENSDART00000178891 |
FQ311928.1
|
|
| chr22_+_1352192 | 0.40 |
ENSDART00000169629
|
znf45l
|
zinc finger 45 like |
| chr13_+_33268657 | 0.40 |
ENSDART00000002095
|
tmem39b
|
transmembrane protein 39B |
| chr3_-_20040636 | 0.40 |
ENSDART00000104118
|
atxn7l3
|
ataxin 7-like 3 |
| chr2_-_10877765 | 0.39 |
ENSDART00000100607
|
cdc7
|
cell division cycle 7 homolog (S. cerevisiae) |
| chr6_-_35046735 | 0.39 |
ENSDART00000143649
|
uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr8_-_2616326 | 0.39 |
ENSDART00000027214
|
slc25a25a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25a |
| chr22_-_10156581 | 0.39 |
ENSDART00000168304
|
rbck1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr11_-_23687158 | 0.39 |
ENSDART00000189599
|
pik3c2b
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr10_-_11261565 | 0.39 |
ENSDART00000146727
|
ptbp3
|
polypyrimidine tract binding protein 3 |
| chr4_+_77943184 | 0.38 |
ENSDART00000159094
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr25_+_5755314 | 0.38 |
ENSDART00000172088
ENSDART00000164208 |
appl2
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
| chr14_-_26425416 | 0.36 |
ENSDART00000088690
|
lman2
|
lectin, mannose-binding 2 |
| chr6_-_50730749 | 0.36 |
ENSDART00000157153
ENSDART00000110441 |
pigu
|
phosphatidylinositol glycan anchor biosynthesis, class U |
| chr18_-_15420168 | 0.36 |
ENSDART00000091466
|
polr3b
|
polymerase (RNA) III (DNA directed) polypeptide B |
| chr7_-_57509447 | 0.36 |
ENSDART00000051973
ENSDART00000147036 |
sirt3
|
sirtuin 3 |
| chr7_-_8738827 | 0.36 |
ENSDART00000172807
ENSDART00000173026 |
si:ch211-1o7.3
|
si:ch211-1o7.3 |
| chr5_-_66823750 | 0.36 |
ENSDART00000041441
ENSDART00000112488 |
stip1
|
stress-induced phosphoprotein 1 |
| chr13_+_4409294 | 0.35 |
ENSDART00000146437
|
si:ch211-130h14.4
|
si:ch211-130h14.4 |
| chr16_+_28994709 | 0.35 |
ENSDART00000088023
|
gon4l
|
gon-4-like (C. elegans) |
| chr23_+_36460239 | 0.34 |
ENSDART00000172441
|
lima1a
|
LIM domain and actin binding 1a |
| chr14_+_32430982 | 0.34 |
ENSDART00000017179
ENSDART00000123382 ENSDART00000075593 |
f9a
|
coagulation factor IXa |
| chr17_+_22577472 | 0.34 |
ENSDART00000045099
|
yipf4
|
Yip1 domain family, member 4 |
| chr11_+_24800156 | 0.33 |
ENSDART00000131976
|
adipor1a
|
adiponectin receptor 1a |
| chr8_-_14239023 | 0.33 |
ENSDART00000090371
|
sin3b
|
SIN3 transcription regulator family member B |
| chr7_-_67248829 | 0.33 |
ENSDART00000192442
|
znf143a
|
zinc finger protein 143a |
| chr20_-_28352352 | 0.33 |
ENSDART00000128806
|
ino80
|
INO80 complex subunit |
| chr17_+_8799451 | 0.33 |
ENSDART00000189814
ENSDART00000191577 |
tonsl
|
tonsoku-like, DNA repair protein |
| chr2_+_36112273 | 0.33 |
ENSDART00000191315
|
traj35
|
T-cell receptor alpha joining 35 |
| chr7_+_19552381 | 0.33 |
ENSDART00000169060
|
si:ch211-212k18.5
|
si:ch211-212k18.5 |
| chr12_+_16087077 | 0.33 |
ENSDART00000141898
|
znf281b
|
zinc finger protein 281b |
| chr16_-_31351419 | 0.33 |
ENSDART00000178298
ENSDART00000018091 |
mroh1
|
maestro heat-like repeat family member 1 |
| chr10_-_42923385 | 0.33 |
ENSDART00000076731
|
ACOT12
|
acyl-CoA thioesterase 12 |
| chr16_-_17345377 | 0.33 |
ENSDART00000143056
|
zyx
|
zyxin |
| chr8_+_11425048 | 0.32 |
ENSDART00000018739
|
tjp2b
|
tight junction protein 2b (zona occludens 2) |
| chr23_-_36446307 | 0.32 |
ENSDART00000136623
|
zgc:174906
|
zgc:174906 |
| chr3_-_26806032 | 0.32 |
ENSDART00000143710
|
pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr19_+_31873308 | 0.32 |
ENSDART00000146560
ENSDART00000133045 |
si:dkeyp-34f6.4
|
si:dkeyp-34f6.4 |
| chr5_-_23843636 | 0.32 |
ENSDART00000193280
|
GBGT1 (1 of many)
|
si:ch211-135f11.5 |
| chr16_+_54209504 | 0.31 |
ENSDART00000020033
|
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr21_-_1644414 | 0.31 |
ENSDART00000105736
ENSDART00000124904 |
zgc:152948
|
zgc:152948 |
| chr22_+_9522971 | 0.31 |
ENSDART00000110048
|
strip1
|
striatin interacting protein 1 |
| chr9_-_9415000 | 0.31 |
ENSDART00000146210
|
si:ch211-214p13.9
|
si:ch211-214p13.9 |
| chr8_+_21437908 | 0.31 |
ENSDART00000142758
|
si:dkey-163f12.10
|
si:dkey-163f12.10 |
| chr23_-_18567088 | 0.31 |
ENSDART00000192371
|
sephs2
|
selenophosphate synthetase 2 |
| chr7_-_51368681 | 0.31 |
ENSDART00000146385
|
arhgap36
|
Rho GTPase activating protein 36 |
| chr3_+_52545014 | 0.30 |
ENSDART00000018908
|
slc27a1a
|
solute carrier family 27 (fatty acid transporter), member 1a |
| chr11_-_6069081 | 0.30 |
ENSDART00000008980
|
babam1
|
BRISC and BRCA1 A complex member 1 |
| chr3_+_431208 | 0.30 |
ENSDART00000154296
ENSDART00000048733 |
si:ch73-308m11.1
si:dkey-167k11.5
|
si:ch73-308m11.1 si:dkey-167k11.5 |
| chr13_-_12602920 | 0.30 |
ENSDART00000102311
|
lrit3b
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 3b |
| chr25_-_27621268 | 0.30 |
ENSDART00000146205
ENSDART00000073511 |
hyal6
|
hyaluronoglucosaminidase 6 |
| chr17_+_30369396 | 0.29 |
ENSDART00000076611
|
greb1
|
growth regulation by estrogen in breast cancer 1 |
| chr4_+_17655872 | 0.29 |
ENSDART00000066999
|
washc3
|
WASH complex subunit 3 |
| chr14_-_1200854 | 0.29 |
ENSDART00000106672
|
arl9
|
ADP-ribosylation factor-like 9 |
| chr5_+_4006837 | 0.28 |
ENSDART00000138862
|
pigw
|
phosphatidylinositol glycan anchor biosynthesis, class W |
| chr22_-_36530902 | 0.28 |
ENSDART00000056188
|
polr2h
|
info polymerase (RNA) II (DNA directed) polypeptide H |
| chr22_+_2769236 | 0.28 |
ENSDART00000141836
|
si:dkey-20i20.10
|
si:dkey-20i20.10 |
| chr7_-_8712148 | 0.28 |
ENSDART00000065488
|
tex261
|
testis expressed 261 |
Network of associatons between targets according to the STRING database.
First level regulatory network of lhx2b+lhx9
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.4 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.5 | 2.3 | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.4 | 2.1 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.4 | 1.7 | GO:0015722 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.3 | 1.2 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.3 | 1.1 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.2 | 0.6 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.2 | 1.1 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.2 | 0.9 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.2 | 0.7 | GO:2000677 | histone H3-K79 methylation(GO:0034729) regulation of transcription regulatory region DNA binding(GO:2000677) |
| 0.2 | 1.6 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.2 | 0.5 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 0.8 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.1 | 0.9 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.1 | 1.3 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 0.3 | GO:0051228 | protein hexamerization(GO:0034214) mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.1 | 0.7 | GO:0072401 | signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) |
| 0.1 | 0.6 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.1 | 0.4 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.1 | 0.4 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.1 | 0.8 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.6 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 0.4 | GO:0048714 | positive regulation of glial cell differentiation(GO:0045687) positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.1 | 0.2 | GO:1904478 | regulation of intestinal absorption(GO:1904478) regulation of intestinal lipid absorption(GO:1904729) |
| 0.1 | 0.3 | GO:1902165 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902165) |
| 0.1 | 0.9 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 | 0.3 | GO:0042766 | nucleosome mobilization(GO:0042766) |
| 0.1 | 0.6 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 | 0.6 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.1 | 0.3 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.1 | 3.3 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 0.4 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.9 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 0.5 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 0.4 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.1 | 0.4 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.1 | 0.6 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.1 | 0.2 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 0.9 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.6 | GO:0031111 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.0 | 0.6 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.0 | 0.6 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.8 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.6 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 2.1 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.2 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.0 | 0.6 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.4 | GO:0070836 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.6 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.6 | GO:0021772 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.6 | GO:0035459 | cargo loading into vesicle(GO:0035459) |
| 0.0 | 0.4 | GO:0031937 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.3 | GO:0070977 | ossification involved in bone maturation(GO:0043931) bone maturation(GO:0070977) |
| 0.0 | 0.4 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.8 | GO:0016233 | telomere capping(GO:0016233) |
| 0.0 | 0.3 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.0 | 0.4 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.6 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.4 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.0 | 0.6 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.1 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 1.1 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 1.4 | GO:0034968 | histone lysine methylation(GO:0034968) |
| 0.0 | 0.4 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.3 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.1 | GO:0097053 | L-kynurenine metabolic process(GO:0097052) L-kynurenine catabolic process(GO:0097053) |
| 0.0 | 0.4 | GO:0098781 | ncRNA transcription(GO:0098781) |
| 0.0 | 1.8 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 1.4 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.4 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.0 | 0.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.4 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.2 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.0 | 0.6 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.1 | GO:0021557 | oculomotor nerve development(GO:0021557) |
| 0.0 | 0.1 | GO:0097241 | hematopoietic stem cell migration to bone marrow(GO:0097241) |
| 0.0 | 0.1 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.0 | 0.2 | GO:0018120 | peptidyl-arginine ADP-ribosylation(GO:0018120) |
| 0.0 | 0.2 | GO:0060754 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.1 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.0 | 0.2 | GO:0050779 | RNA destabilization(GO:0050779) mRNA destabilization(GO:0061157) |
| 0.0 | 0.3 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 4.1 | GO:0006486 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 0.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.4 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.2 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.1 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.3 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.2 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.0 | 0.2 | GO:0055117 | regulation of cardiac muscle contraction(GO:0055117) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.5 | 1.6 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.3 | 1.0 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.2 | 0.6 | GO:1990879 | CST complex(GO:1990879) |
| 0.1 | 1.1 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.1 | 0.5 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 0.8 | GO:0071012 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.1 | 0.7 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 0.6 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 0.3 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 0.4 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 0.3 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 2.0 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 0.6 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 0.3 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 1.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 0.2 | GO:0008247 | 1-alkyl-2-acetylglycerophosphocholine esterase complex(GO:0008247) |
| 0.1 | 0.4 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.7 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.4 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.8 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.2 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.6 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 1.1 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.6 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.8 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.7 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.4 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.6 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.3 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 1.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.3 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.6 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.6 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.6 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 2.4 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.4 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.4 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.2 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.4 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 0.1 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.8 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.3 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.2 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.6 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.8 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 1.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.7 | GO:0005905 | clathrin-coated pit(GO:0005905) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.4 | 1.7 | GO:0015432 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.2 | 1.6 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.2 | 1.5 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.2 | 0.5 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.2 | 0.7 | GO:0031151 | histone methyltransferase activity (H3-K79 specific)(GO:0031151) |
| 0.2 | 0.5 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.6 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 2.0 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 1.6 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 1.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 0.9 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 1.2 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 0.3 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.1 | 0.4 | GO:1902388 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.1 | 0.4 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.1 | 1.3 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 0.2 | GO:0003994 | aconitate hydratase activity(GO:0003994) |
| 0.1 | 0.3 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 0.6 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.1 | 0.5 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 0.7 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.6 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 0.6 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 1.1 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.3 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 3.4 | GO:0016409 | palmitoyltransferase activity(GO:0016409) |
| 0.1 | 1.0 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.1 | 0.6 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.5 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.6 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.6 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.8 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.3 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.6 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.2 | GO:0015105 | arsenite transmembrane transporter activity(GO:0015105) |
| 0.0 | 0.8 | GO:0017136 | histone deacetylase activity(GO:0004407) NAD-dependent histone deacetylase activity(GO:0017136) |
| 0.0 | 0.3 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 1.7 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 2.1 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.2 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.1 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.4 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.4 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.4 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.4 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.3 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.2 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.4 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 4.0 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 0.2 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.8 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.9 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.2 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.2 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 1.0 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.4 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 0.4 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.3 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 2.1 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.1 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 1.1 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.0 | 0.0 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.4 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.6 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.2 | GO:0042162 | telomeric DNA binding(GO:0042162) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.4 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 1.0 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.7 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 1.9 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.9 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.6 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 0.5 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.4 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.8 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 2.1 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.7 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.3 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.4 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 2.1 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 0.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.6 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.7 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.6 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.4 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.8 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.4 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.8 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 1.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.9 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.3 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 0.5 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 0.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.2 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.2 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.4 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.4 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.4 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |