Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
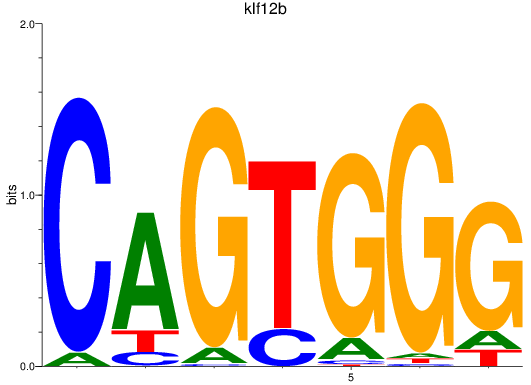
Results for klf12b
Z-value: 1.16
Transcription factors associated with klf12b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
klf12b
|
ENSDARG00000032197 | Kruppel-like factor 12b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| klf12b | dr11_v1_chr9_+_30720048_30720048 | 0.82 | 7.3e-03 | Click! |
Activity profile of klf12b motif
Sorted Z-values of klf12b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_13742899 | 2.17 |
ENSDART00000104722
|
cdk5r2a
|
cyclin-dependent kinase 5, regulatory subunit 2a (p39) |
| chr21_+_25187210 | 2.07 |
ENSDART00000101147
ENSDART00000167528 |
si:dkey-183i3.5
|
si:dkey-183i3.5 |
| chr5_-_67471375 | 1.47 |
ENSDART00000147009
|
si:dkey-251i10.2
|
si:dkey-251i10.2 |
| chr13_-_39947335 | 1.28 |
ENSDART00000056996
|
sfrp5
|
secreted frizzled-related protein 5 |
| chr13_-_39159810 | 1.23 |
ENSDART00000131508
|
col9a1b
|
collagen, type IX, alpha 1b |
| chr19_-_4010263 | 1.20 |
ENSDART00000159605
ENSDART00000165541 |
map7d1b
|
MAP7 domain containing 1b |
| chr13_+_13681681 | 1.19 |
ENSDART00000057825
|
cfd
|
complement factor D (adipsin) |
| chr20_-_22484621 | 1.16 |
ENSDART00000063601
|
gsx2
|
GS homeobox 2 |
| chr15_+_1397811 | 1.12 |
ENSDART00000102125
|
schip1
|
schwannomin interacting protein 1 |
| chr12_+_42574148 | 1.10 |
ENSDART00000157855
|
ebf3a
|
early B cell factor 3a |
| chr7_-_43840680 | 1.09 |
ENSDART00000002279
|
cdh11
|
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chr6_+_24817852 | 1.07 |
ENSDART00000165609
|
barhl2
|
BarH-like homeobox 2 |
| chr17_+_15433518 | 1.05 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr25_-_31423493 | 1.02 |
ENSDART00000027661
|
myod1
|
myogenic differentiation 1 |
| chr23_+_35714574 | 0.91 |
ENSDART00000164616
|
tuba1c
|
tubulin, alpha 1c |
| chr15_-_37846047 | 0.90 |
ENSDART00000184837
|
hsc70
|
heat shock cognate 70 |
| chr20_+_15015557 | 0.87 |
ENSDART00000039345
|
myoc
|
myocilin |
| chr2_+_47582681 | 0.86 |
ENSDART00000187579
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr16_+_23947196 | 0.84 |
ENSDART00000103190
ENSDART00000132961 ENSDART00000147690 ENSDART00000142168 |
apoa4b.2
|
apolipoprotein A-IV b, tandem duplicate 2 |
| chr5_-_28679135 | 0.81 |
ENSDART00000193585
|
tnc
|
tenascin C |
| chr13_-_31470439 | 0.80 |
ENSDART00000076574
|
rtn1a
|
reticulon 1a |
| chr4_-_16353733 | 0.80 |
ENSDART00000186785
|
lum
|
lumican |
| chr16_+_20926673 | 0.76 |
ENSDART00000009827
|
hoxa2b
|
homeobox A2b |
| chr14_-_45206999 | 0.76 |
ENSDART00000110191
|
shisa3
|
shisa family member 3 |
| chr3_+_15505275 | 0.75 |
ENSDART00000141714
|
nupr1
|
nuclear protein 1 |
| chr16_-_2414063 | 0.75 |
ENSDART00000073621
|
zgc:152945
|
zgc:152945 |
| chr25_+_15647750 | 0.75 |
ENSDART00000137375
|
spon1b
|
spondin 1b |
| chr11_+_23957440 | 0.74 |
ENSDART00000190721
|
cntn2
|
contactin 2 |
| chr13_-_11536951 | 0.74 |
ENSDART00000018155
|
adss
|
adenylosuccinate synthase |
| chr7_-_30082931 | 0.74 |
ENSDART00000075600
|
tspan3b
|
tetraspanin 3b |
| chr1_-_44434707 | 0.73 |
ENSDART00000110148
|
cryba1l2
|
crystallin, beta A1, like 2 |
| chr13_+_25449681 | 0.72 |
ENSDART00000101328
|
atoh7
|
atonal bHLH transcription factor 7 |
| chr4_+_6643421 | 0.71 |
ENSDART00000099462
|
gpr85
|
G protein-coupled receptor 85 |
| chr19_-_26863626 | 0.71 |
ENSDART00000145568
|
prrt1
|
proline-rich transmembrane protein 1 |
| chr17_-_15546862 | 0.71 |
ENSDART00000091021
|
col10a1a
|
collagen, type X, alpha 1a |
| chr18_+_18104235 | 0.69 |
ENSDART00000145342
|
cbln1
|
cerebellin 1 precursor |
| chr2_-_29485408 | 0.69 |
ENSDART00000013411
|
cahz
|
carbonic anhydrase |
| chr22_-_2886937 | 0.68 |
ENSDART00000063533
|
aqp12
|
aquaporin 12 |
| chr17_+_10242166 | 0.68 |
ENSDART00000170420
|
clec14a
|
C-type lectin domain containing 14A |
| chr2_+_27010439 | 0.67 |
ENSDART00000030547
|
cdh7a
|
cadherin 7a |
| chr2_+_33368414 | 0.67 |
ENSDART00000077462
|
slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr23_-_39636195 | 0.65 |
ENSDART00000144439
|
vwa1
|
von Willebrand factor A domain containing 1 |
| chr8_+_26565512 | 0.65 |
ENSDART00000140980
|
sema3bl
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3bl |
| chr17_-_49978986 | 0.65 |
ENSDART00000154728
|
col12a1a
|
collagen, type XII, alpha 1a |
| chr20_-_26001288 | 0.65 |
ENSDART00000136518
ENSDART00000063177 |
capn3b
|
calpain 3b |
| chr5_-_65782783 | 0.65 |
ENSDART00000130888
ENSDART00000050855 |
notch1b
|
notch 1b |
| chr20_+_1412193 | 0.64 |
ENSDART00000064419
|
leg1.1
|
liver-enriched gene 1, tandem duplicate 1 |
| chr6_+_49881864 | 0.64 |
ENSDART00000075040
|
tubb1
|
tubulin, beta 1 class VI |
| chr20_-_44496245 | 0.64 |
ENSDART00000012229
|
fkbp1b
|
FK506 binding protein 1b |
| chr12_+_13164706 | 0.64 |
ENSDART00000112565
|
paqr4b
|
progestin and adipoQ receptor family member IVb |
| chr24_+_7637522 | 0.64 |
ENSDART00000082467
|
cavin1b
|
caveolae associated protein 1b |
| chr2_+_22042745 | 0.64 |
ENSDART00000132039
|
tox
|
thymocyte selection-associated high mobility group box |
| chr17_+_18117358 | 0.64 |
ENSDART00000144894
|
bcl11ba
|
B cell CLL/lymphoma 11Ba |
| chr15_+_37197494 | 0.63 |
ENSDART00000166203
|
aplp1
|
amyloid beta (A4) precursor-like protein 1 |
| chr17_-_52643970 | 0.62 |
ENSDART00000190594
|
spred1
|
sprouty-related, EVH1 domain containing 1 |
| chr21_+_29077509 | 0.62 |
ENSDART00000128561
|
ecscr
|
endothelial cell surface expressed chemotaxis and apoptosis regulator |
| chr9_-_1939232 | 0.61 |
ENSDART00000146131
|
hoxd3a
|
homeobox D3a |
| chr9_-_35155089 | 0.61 |
ENSDART00000077901
|
appb
|
amyloid beta (A4) precursor protein b |
| chr2_-_65529 | 0.60 |
ENSDART00000192876
|
zgc:153913
|
zgc:153913 |
| chr4_-_4834617 | 0.60 |
ENSDART00000141539
|
coa6
|
cytochrome c oxidase assembly factor 6 |
| chr22_-_23666504 | 0.59 |
ENSDART00000158665
|
cfh
|
complement factor H |
| chr4_-_1324141 | 0.59 |
ENSDART00000180720
|
ptn
|
pleiotrophin |
| chr13_+_19322686 | 0.59 |
ENSDART00000058036
|
emx2
|
empty spiracles homeobox 2 |
| chr20_-_47704973 | 0.59 |
ENSDART00000174808
|
tfap2b
|
transcription factor AP-2 beta |
| chr18_+_9171778 | 0.58 |
ENSDART00000101192
|
sema3d
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D |
| chr23_+_22658700 | 0.58 |
ENSDART00000192248
|
eno1a
|
enolase 1a, (alpha) |
| chr23_+_8797143 | 0.57 |
ENSDART00000132992
|
sox18
|
SRY (sex determining region Y)-box 18 |
| chr11_+_36989696 | 0.56 |
ENSDART00000045888
|
tkta
|
transketolase a |
| chr24_+_24461558 | 0.55 |
ENSDART00000182424
|
bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr6_-_49215790 | 0.55 |
ENSDART00000103376
ENSDART00000132347 ENSDART00000132131 ENSDART00000143252 ENSDART00000065033 |
ngfb
|
nerve growth factor b (beta polypeptide) |
| chr18_+_20494413 | 0.55 |
ENSDART00000060295
|
rapsn
|
receptor-associated protein of the synapse, 43kD |
| chr22_-_26353916 | 0.54 |
ENSDART00000077958
|
capn2b
|
calpain 2, (m/II) large subunit b |
| chr3_+_28953274 | 0.54 |
ENSDART00000133528
ENSDART00000103602 |
lgals2a
|
lectin, galactoside-binding, soluble, 2a |
| chr14_-_36863432 | 0.54 |
ENSDART00000158052
|
rnf130
|
ring finger protein 130 |
| chr4_+_5741733 | 0.54 |
ENSDART00000110243
|
pou3f2a
|
POU class 3 homeobox 2a |
| chr19_+_27479838 | 0.53 |
ENSDART00000103922
|
atat1
|
alpha tubulin acetyltransferase 1 |
| chr8_-_14144707 | 0.53 |
ENSDART00000148061
|
si:dkey-6n6.1
|
si:dkey-6n6.1 |
| chr14_+_32022272 | 0.53 |
ENSDART00000105760
|
zic6
|
zic family member 6 |
| chr12_+_16233077 | 0.53 |
ENSDART00000152409
|
mpp3b
|
membrane protein, palmitoylated 3b (MAGUK p55 subfamily member 3) |
| chr12_+_24344963 | 0.53 |
ENSDART00000191648
ENSDART00000183180 ENSDART00000088178 ENSDART00000189696 |
nrxn1a
|
neurexin 1a |
| chr17_+_18117029 | 0.53 |
ENSDART00000154646
ENSDART00000179739 |
bcl11ba
|
B cell CLL/lymphoma 11Ba |
| chr3_+_23738215 | 0.52 |
ENSDART00000143981
|
hoxb3a
|
homeobox B3a |
| chr20_-_19422496 | 0.52 |
ENSDART00000143658
|
si:ch211-278j3.3
|
si:ch211-278j3.3 |
| chr16_-_12809873 | 0.52 |
ENSDART00000146997
ENSDART00000178291 ENSDART00000007842 |
isoc2
|
isochorismatase domain containing 2 |
| chr19_+_37458610 | 0.52 |
ENSDART00000103151
|
dlgap3
|
discs, large (Drosophila) homolog-associated protein 3 |
| chr4_+_15954293 | 0.51 |
ENSDART00000132695
|
si:dkey-117n7.4
|
si:dkey-117n7.4 |
| chr17_+_51627209 | 0.51 |
ENSDART00000056886
|
zgc:113142
|
zgc:113142 |
| chr16_-_12173554 | 0.50 |
ENSDART00000110567
ENSDART00000155935 |
clstn3
|
calsyntenin 3 |
| chr11_-_42554290 | 0.50 |
ENSDART00000130573
|
atp6ap1la
|
ATPase H+ transporting accessory protein 1 like a |
| chr22_-_5006801 | 0.49 |
ENSDART00000106166
|
rx1
|
retinal homeobox gene 1 |
| chr11_+_23933016 | 0.49 |
ENSDART00000000486
|
cntn2
|
contactin 2 |
| chr3_-_30685401 | 0.49 |
ENSDART00000151097
|
si:ch211-51c14.1
|
si:ch211-51c14.1 |
| chr9_-_31278048 | 0.48 |
ENSDART00000022204
|
zic5
|
zic family member 5 (odd-paired homolog, Drosophila) |
| chr18_-_22753637 | 0.48 |
ENSDART00000181589
ENSDART00000009912 |
hsf4
|
heat shock transcription factor 4 |
| chr5_+_54585431 | 0.48 |
ENSDART00000171225
|
npr2
|
natriuretic peptide receptor 2 |
| chr8_-_9118958 | 0.48 |
ENSDART00000037922
|
slc6a8
|
solute carrier family 6 (neurotransmitter transporter), member 8 |
| chr24_+_7637264 | 0.47 |
ENSDART00000124409
|
cavin1b
|
caveolae associated protein 1b |
| chr22_+_12431608 | 0.47 |
ENSDART00000108609
|
rnd3a
|
Rho family GTPase 3a |
| chr3_-_38692920 | 0.47 |
ENSDART00000155042
|
mpp3a
|
membrane protein, palmitoylated 3a (MAGUK p55 subfamily member 3) |
| chr4_+_19535946 | 0.46 |
ENSDART00000192342
ENSDART00000183740 ENSDART00000180812 ENSDART00000180017 |
lrrc4.1
|
leucine rich repeat containing 4.1 |
| chr14_-_30704075 | 0.46 |
ENSDART00000134098
|
efemp2a
|
EGF containing fibulin extracellular matrix protein 2a |
| chr10_+_22381802 | 0.46 |
ENSDART00000112484
|
nlgn2b
|
neuroligin 2b |
| chr7_+_44713135 | 0.45 |
ENSDART00000170721
|
si:dkey-56m19.5
|
si:dkey-56m19.5 |
| chr22_-_10482253 | 0.45 |
ENSDART00000143164
|
aspn
|
asporin (LRR class 1) |
| chr22_-_600016 | 0.45 |
ENSDART00000086434
|
tmcc2
|
transmembrane and coiled-coil domain family 2 |
| chr6_-_20875111 | 0.45 |
ENSDART00000115118
ENSDART00000159916 |
tns1a
|
tensin 1a |
| chr15_+_15415623 | 0.44 |
ENSDART00000127436
|
zgc:92630
|
zgc:92630 |
| chr14_-_39074539 | 0.44 |
ENSDART00000030509
|
glra4a
|
glycine receptor, alpha 4a |
| chr25_-_15040369 | 0.44 |
ENSDART00000159342
ENSDART00000166490 |
pax6a
|
paired box 6a |
| chr4_-_4834347 | 0.44 |
ENSDART00000141803
|
coa6
|
cytochrome c oxidase assembly factor 6 |
| chr22_-_11626014 | 0.44 |
ENSDART00000063133
ENSDART00000160085 |
gcga
|
glucagon a |
| chr20_+_26349002 | 0.44 |
ENSDART00000152842
|
syne1a
|
spectrin repeat containing, nuclear envelope 1a |
| chr1_-_35653560 | 0.44 |
ENSDART00000142154
|
frem3
|
Fras1 related extracellular matrix 3 |
| chr17_-_36896560 | 0.44 |
ENSDART00000045287
|
mapre3a
|
microtubule-associated protein, RP/EB family, member 3a |
| chr4_-_6809323 | 0.44 |
ENSDART00000099467
|
ifrd1
|
interferon-related developmental regulator 1 |
| chr1_+_27112479 | 0.44 |
ENSDART00000128671
|
bnc2
|
basonuclin 2 |
| chr11_-_37509001 | 0.43 |
ENSDART00000109753
|
bsnb
|
bassoon (presynaptic cytomatrix protein) b |
| chr4_-_18201622 | 0.43 |
ENSDART00000133509
|
anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr14_+_36223097 | 0.43 |
ENSDART00000186872
|
pitx2
|
paired-like homeodomain 2 |
| chr5_+_52039067 | 0.43 |
ENSDART00000143276
|
setbp1
|
SET binding protein 1 |
| chr7_+_26224211 | 0.43 |
ENSDART00000173999
|
vgf
|
VGF nerve growth factor inducible |
| chr10_+_21776911 | 0.42 |
ENSDART00000163077
ENSDART00000186093 |
pcdh1g22
|
protocadherin 1 gamma 22 |
| chr18_+_17583479 | 0.41 |
ENSDART00000186977
ENSDART00000010998 |
slc12a3
|
solute carrier family 12 (sodium/chloride transporter), member 3 |
| chr1_+_59073203 | 0.41 |
ENSDART00000149937
ENSDART00000162201 |
MFAP4 (1 of many)
zgc:173915
|
si:zfos-2330d3.3 zgc:173915 |
| chr7_-_57933736 | 0.41 |
ENSDART00000142580
|
ank2b
|
ankyrin 2b, neuronal |
| chr12_+_2381213 | 0.41 |
ENSDART00000188007
|
LO018238.1
|
|
| chr25_-_30357027 | 0.41 |
ENSDART00000171137
|
pdia3
|
protein disulfide isomerase family A, member 3 |
| chr14_-_26377044 | 0.41 |
ENSDART00000022236
|
emx3
|
empty spiracles homeobox 3 |
| chr15_+_32821392 | 0.41 |
ENSDART00000158272
|
dclk1b
|
doublecortin-like kinase 1b |
| chr8_+_42998944 | 0.41 |
ENSDART00000048819
|
rassf2a
|
Ras association (RalGDS/AF-6) domain family member 2a |
| chr23_-_23401305 | 0.40 |
ENSDART00000078936
|
her9
|
hairy-related 9 |
| chr9_-_21970067 | 0.40 |
ENSDART00000009920
|
lmo7a
|
LIM domain 7a |
| chr21_+_17768174 | 0.40 |
ENSDART00000141380
|
rxraa
|
retinoid X receptor, alpha a |
| chr5_-_55395964 | 0.40 |
ENSDART00000145791
|
prune2
|
prune homolog 2 (Drosophila) |
| chr24_+_18948665 | 0.39 |
ENSDART00000106186
|
prex2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr13_+_31108334 | 0.39 |
ENSDART00000142245
|
arhgap22
|
Rho GTPase activating protein 22 |
| chr8_+_31119548 | 0.39 |
ENSDART00000136578
|
syn1
|
synapsin I |
| chr21_-_44104600 | 0.39 |
ENSDART00000044599
|
oatx
|
organic anion transporter X |
| chr16_-_44512882 | 0.39 |
ENSDART00000191241
|
CR925804.2
|
|
| chr23_-_8373676 | 0.38 |
ENSDART00000105135
ENSDART00000158531 |
oprl1
|
opiate receptor-like 1 |
| chr8_-_53108207 | 0.38 |
ENSDART00000111023
|
b3galt4
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 4 |
| chr12_+_13905286 | 0.38 |
ENSDART00000147186
|
fkbp10b
|
FK506 binding protein 10b |
| chr18_-_45617146 | 0.38 |
ENSDART00000146543
|
wt1b
|
wilms tumor 1b |
| chr12_-_212843 | 0.38 |
ENSDART00000083574
|
CABZ01102039.1
|
|
| chr5_-_23317477 | 0.37 |
ENSDART00000090171
|
nlgn3b
|
neuroligin 3b |
| chr15_-_20916251 | 0.37 |
ENSDART00000134053
|
usp2a
|
ubiquitin specific peptidase 2a |
| chr12_+_9817440 | 0.37 |
ENSDART00000137081
ENSDART00000123712 |
rundc3ab
|
RUN domain containing 3Ab |
| chr23_+_44883805 | 0.37 |
ENSDART00000182805
|
si:ch73-361h17.1
|
si:ch73-361h17.1 |
| chr1_+_59090972 | 0.37 |
ENSDART00000171497
|
mfap4
|
microfibril associated protein 4 |
| chr10_-_43771447 | 0.37 |
ENSDART00000052307
|
arrdc3b
|
arrestin domain containing 3b |
| chr7_+_11197940 | 0.37 |
ENSDART00000081346
|
cemip
|
cell migration inducing protein, hyaluronan binding |
| chr3_-_32320537 | 0.37 |
ENSDART00000113550
ENSDART00000168483 |
si:dkey-16p21.7
|
si:dkey-16p21.7 |
| chr1_+_59090743 | 0.36 |
ENSDART00000100199
|
mfap4
|
microfibril associated protein 4 |
| chr2_+_22694382 | 0.36 |
ENSDART00000139196
|
kif1ab
|
kinesin family member 1Ab |
| chr15_-_8517555 | 0.36 |
ENSDART00000140213
|
npas1
|
neuronal PAS domain protein 1 |
| chr2_-_10703621 | 0.36 |
ENSDART00000005944
|
rpl5a
|
ribosomal protein L5a |
| chr21_-_12272543 | 0.36 |
ENSDART00000081510
ENSDART00000151297 |
celf4
|
CUGBP, Elav-like family member 4 |
| chr5_-_38506981 | 0.35 |
ENSDART00000097822
|
atp1b2b
|
ATPase Na+/K+ transporting subunit beta 2b |
| chr20_+_539852 | 0.35 |
ENSDART00000185994
|
dse
|
dermatan sulfate epimerase |
| chr22_+_5727753 | 0.35 |
ENSDART00000063484
|
si:dkey-222f2.1
|
si:dkey-222f2.1 |
| chr2_-_20599315 | 0.35 |
ENSDART00000114199
|
si:ch211-267e7.3
|
si:ch211-267e7.3 |
| chr17_-_24684687 | 0.34 |
ENSDART00000105457
|
morn2
|
MORN repeat containing 2 |
| chr8_-_14050758 | 0.34 |
ENSDART00000133922
|
atp2b3a
|
ATPase plasma membrane Ca2+ transporting 3a |
| chr13_-_15982707 | 0.34 |
ENSDART00000186911
ENSDART00000181072 |
ikzf1
|
IKAROS family zinc finger 1 (Ikaros) |
| chr7_+_33314925 | 0.34 |
ENSDART00000148590
|
coro2ba
|
coronin, actin binding protein, 2Ba |
| chr15_-_32595982 | 0.34 |
ENSDART00000171808
|
frem2b
|
Fras1 related extracellular matrix protein 2b |
| chr21_+_5993188 | 0.34 |
ENSDART00000048399
|
slc4a4b
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4b |
| chr13_-_42560662 | 0.34 |
ENSDART00000124898
|
CR792417.1
|
|
| chr25_-_13105310 | 0.33 |
ENSDART00000172269
|
smpd3
|
sphingomyelin phosphodiesterase 3, neutral |
| chr1_-_59104145 | 0.33 |
ENSDART00000132495
ENSDART00000152457 |
MFAP4 (1 of many)
si:zfos-2330d3.7
|
si:zfos-2330d3.1 si:zfos-2330d3.7 |
| chr22_+_696931 | 0.33 |
ENSDART00000149712
ENSDART00000009756 |
gpr37l1a
|
G protein-coupled receptor 37 like 1a |
| chr18_+_13203831 | 0.33 |
ENSDART00000032151
|
cotl1
|
coactosin-like F-actin binding protein 1 |
| chr15_-_12319065 | 0.33 |
ENSDART00000162973
ENSDART00000170543 |
fxyd6
|
FXYD domain containing ion transport regulator 6 |
| chr22_+_11867639 | 0.32 |
ENSDART00000144677
|
mras
|
muscle RAS oncogene homolog |
| chr3_-_22829710 | 0.32 |
ENSDART00000055659
|
cyb561
|
cytochrome b561 |
| chr19_-_44089509 | 0.32 |
ENSDART00000189136
|
rad21b
|
RAD21 cohesin complex component b |
| chr1_+_20084389 | 0.32 |
ENSDART00000140263
|
prss12
|
protease, serine, 12 (neurotrypsin, motopsin) |
| chr16_+_12517535 | 0.31 |
ENSDART00000155407
|
rasip1
|
Ras interacting protein 1 |
| chr22_-_26323893 | 0.31 |
ENSDART00000105099
|
capn1b
|
calpain 1, (mu/I) large subunit b |
| chr17_+_23146976 | 0.31 |
ENSDART00000114212
|
rasgrp3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr5_-_22501663 | 0.31 |
ENSDART00000133174
|
si:dkey-27p18.5
|
si:dkey-27p18.5 |
| chr4_-_9557186 | 0.31 |
ENSDART00000150569
|
shank3b
|
SH3 and multiple ankyrin repeat domains 3b |
| chr3_-_57737913 | 0.31 |
ENSDART00000113309
|
LGALS3BP (1 of many)
|
zgc:112492 |
| chr9_-_32753535 | 0.31 |
ENSDART00000060006
|
olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr4_+_20263097 | 0.31 |
ENSDART00000138820
|
lrtm2a
|
leucine-rich repeats and transmembrane domains 2a |
| chr15_+_34062460 | 0.31 |
ENSDART00000164654
|
si:dkey-30e9.6
|
si:dkey-30e9.6 |
| chr18_-_15373620 | 0.30 |
ENSDART00000031752
|
rfx4
|
regulatory factor X, 4 |
| chr9_-_32604414 | 0.30 |
ENSDART00000088876
ENSDART00000166502 |
satb2
|
SATB homeobox 2 |
| chr9_+_25832329 | 0.30 |
ENSDART00000130059
|
zeb2a
|
zinc finger E-box binding homeobox 2a |
| chr7_+_54617060 | 0.30 |
ENSDART00000158898
|
fgf4
|
fibroblast growth factor 4 |
| chr9_-_12736089 | 0.30 |
ENSDART00000088042
|
myo10l3
|
myosin X-like 3 |
| chr12_+_14676349 | 0.30 |
ENSDART00000143401
|
becn1
|
beclin 1, autophagy related |
| chr10_-_20637098 | 0.30 |
ENSDART00000080391
|
sprn2
|
shadow of prion protein 2 |
| chr1_+_59073436 | 0.30 |
ENSDART00000161642
|
MFAP4 (1 of many)
|
si:zfos-2330d3.3 |
| chr18_-_15771551 | 0.30 |
ENSDART00000130931
ENSDART00000154079 |
si:ch211-219a15.3
|
si:ch211-219a15.3 |
| chr25_-_13842618 | 0.30 |
ENSDART00000160258
|
mapk8ip1a
|
mitogen-activated protein kinase 8 interacting protein 1a |
| chr9_-_21067971 | 0.30 |
ENSDART00000004333
|
tbx15
|
T-box 15 |
| chr22_+_5103349 | 0.30 |
ENSDART00000083474
|
atcaya
|
ataxia, cerebellar, Cayman type a |
| chr14_-_1998501 | 0.30 |
ENSDART00000189052
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr5_-_37252111 | 0.29 |
ENSDART00000185110
|
lrch2
|
leucine-rich repeats and calponin homology (CH) domain containing 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of klf12b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.4 | 1.2 | GO:0048677 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.4 | 1.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.3 | 1.0 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.3 | 0.8 | GO:0014814 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) |
| 0.2 | 0.2 | GO:0019323 | pentose catabolic process(GO:0019323) |
| 0.2 | 0.7 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.2 | 1.1 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.2 | 0.7 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.2 | 0.7 | GO:0060092 | regulation of synaptic transmission, glycinergic(GO:0060092) |
| 0.2 | 1.0 | GO:0010801 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.2 | 0.5 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.1 | 0.6 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.1 | 1.1 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.1 | 2.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.6 | GO:0007508 | larval development(GO:0002164) larval heart development(GO:0007508) |
| 0.1 | 0.6 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.1 | 0.3 | GO:0043416 | regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.1 | 1.0 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.1 | 0.4 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.1 | 0.5 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 0.8 | GO:0097104 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.1 | 0.4 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.4 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.1 | 0.2 | GO:0003250 | cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) endocardial cell development(GO:0060958) cell proliferation involved in heart valve development(GO:2000793) |
| 0.1 | 0.3 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.1 | 0.6 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.1 | 0.6 | GO:1990402 | embryonic liver development(GO:1990402) |
| 0.1 | 0.5 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.1 | 0.3 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.1 | 1.1 | GO:0001952 | regulation of cell-matrix adhesion(GO:0001952) |
| 0.1 | 0.2 | GO:0002369 | T cell cytokine production(GO:0002369) |
| 0.1 | 0.7 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.1 | 0.6 | GO:0060118 | vestibular receptor cell differentiation(GO:0060114) vestibular receptor cell development(GO:0060118) |
| 0.1 | 0.2 | GO:2000434 | regulation of protein neddylation(GO:2000434) |
| 0.1 | 0.4 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.1 | 0.6 | GO:0035479 | angioblast cell migration from lateral mesoderm to midline(GO:0035479) |
| 0.1 | 0.9 | GO:0042026 | protein refolding(GO:0042026) |
| 0.1 | 2.2 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.1 | 0.6 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.1 | 0.5 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.2 | GO:0043455 | regulation of secondary metabolic process(GO:0043455) |
| 0.1 | 0.2 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.4 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 1.8 | GO:0034332 | adherens junction organization(GO:0034332) |
| 0.0 | 0.5 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.3 | GO:2000463 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.0 | 0.9 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.3 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.0 | 0.4 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.0 | 0.4 | GO:0039023 | pronephric duct morphogenesis(GO:0039023) nephric duct morphogenesis(GO:0072178) |
| 0.0 | 0.3 | GO:0090303 | positive regulation of wound healing(GO:0090303) |
| 0.0 | 0.7 | GO:0048679 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.0 | 0.1 | GO:0045938 | positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 0.0 | 0.2 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.1 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.2 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.1 | GO:0001778 | plasma membrane repair(GO:0001778) chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 0.0 | 0.1 | GO:0048389 | intermediate mesoderm development(GO:0048389) |
| 0.0 | 0.3 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.2 | GO:0008584 | male gonad development(GO:0008584) water homeostasis(GO:0030104) |
| 0.0 | 0.1 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.0 | 0.3 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.0 | 0.5 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.0 | 0.5 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.4 | GO:0019233 | sensory perception of pain(GO:0019233) |
| 0.0 | 0.2 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.4 | GO:0001964 | startle response(GO:0001964) |
| 0.0 | 0.3 | GO:0000305 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.4 | GO:0031100 | organ regeneration(GO:0031100) |
| 0.0 | 0.9 | GO:1903670 | regulation of sprouting angiogenesis(GO:1903670) |
| 0.0 | 0.4 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.0 | 0.5 | GO:0060038 | cardiac muscle cell proliferation(GO:0060038) |
| 0.0 | 0.2 | GO:0002551 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.9 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.2 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 1.2 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.3 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.5 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.1 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.0 | 0.2 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.0 | 0.1 | GO:2000812 | regulation of barbed-end actin filament capping(GO:2000812) |
| 0.0 | 0.5 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.6 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.0 | 0.4 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.3 | GO:0033077 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.0 | 0.1 | GO:0031174 | lifelong otolith mineralization(GO:0031174) |
| 0.0 | 0.6 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.3 | GO:1900107 | regulation of nodal signaling pathway(GO:1900107) |
| 0.0 | 0.1 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.0 | 0.1 | GO:0034164 | regulation of toll-like receptor 9 signaling pathway(GO:0034163) negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.0 | 0.2 | GO:0061337 | cardiac conduction(GO:0061337) |
| 0.0 | 0.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.6 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.1 | GO:0006543 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.0 | 0.3 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.6 | GO:0060840 | artery development(GO:0060840) |
| 0.0 | 0.4 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.2 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.0 | 0.4 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.3 | GO:0007418 | ventral midline development(GO:0007418) |
| 0.0 | 0.4 | GO:0085029 | extracellular matrix assembly(GO:0085029) |
| 0.0 | 0.2 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.4 | GO:0039020 | pronephric nephron tubule development(GO:0039020) |
| 0.0 | 0.2 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 1.0 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.7 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.4 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.1 | GO:0072337 | modified amino acid transport(GO:0072337) |
| 0.0 | 0.1 | GO:0045580 | regulation of T cell differentiation(GO:0045580) |
| 0.0 | 0.1 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 2.3 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.7 | GO:0046328 | regulation of JNK cascade(GO:0046328) |
| 0.0 | 0.4 | GO:0050768 | negative regulation of neurogenesis(GO:0050768) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.3 | GO:0019835 | cytolysis(GO:0019835) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.2 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.2 | 0.5 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 0.5 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.1 | 0.6 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.3 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.1 | 0.3 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 0.3 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 0.3 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.2 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.3 | GO:0034990 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.4 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.0 | 0.4 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 3.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.6 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.0 | 0.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 1.1 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.4 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.1 | GO:0071540 | eukaryotic translation initiation factor 3 complex, eIF3e(GO:0071540) |
| 0.0 | 0.2 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.3 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.3 | GO:0033270 | paranode region of axon(GO:0033270) juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.5 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.4 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.3 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 1.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.9 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.1 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 2.8 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.5 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.2 | 0.7 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.2 | 0.5 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.2 | 2.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 1.0 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.5 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.3 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.1 | 0.6 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.1 | 0.4 | GO:0031769 | glucagon receptor binding(GO:0031769) |
| 0.1 | 1.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.7 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 0.4 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.1 | 0.4 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 0.6 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 1.0 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 0.3 | GO:0032034 | myosin head/neck binding(GO:0032028) myosin II head/neck binding(GO:0032034) myosin II heavy chain binding(GO:0032038) |
| 0.1 | 0.9 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.2 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 0.5 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 0.6 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.1 | 0.3 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.1 | 0.5 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.2 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.1 | 1.6 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.7 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 1.7 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.4 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.3 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.4 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.2 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.0 | 0.4 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.5 | GO:0005165 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.0 | 0.3 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 1.3 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.2 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.1 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.2 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0051800 | inositol-1,3,4,5-tetrakisphosphate 3-phosphatase activity(GO:0051717) phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.0 | 0.4 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.3 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 1.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.2 | GO:0031779 | melanocortin receptor binding(GO:0031779) |
| 0.0 | 0.4 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.2 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 2.1 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 1.0 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.6 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.3 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.6 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.0 | 0.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.2 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 1.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.5 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.0 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.2 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.2 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.2 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 1.1 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.1 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.3 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.0 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 1.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.8 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.5 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.4 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.3 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 2.0 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.1 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.2 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.1 | 1.5 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 0.4 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.1 | 0.8 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 0.9 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 0.3 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.1 | 0.7 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 1.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.1 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.9 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.3 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.2 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.4 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.2 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.2 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.3 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.4 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |