Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
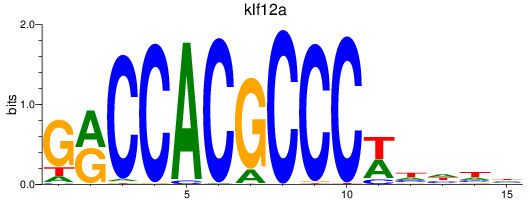
Results for klf12a
Z-value: 0.60
Transcription factors associated with klf12a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
klf12a
|
ENSDARG00000015312 | Kruppel-like factor 12a |
|
klf12a
|
ENSDARG00000115152 | Kruppel-like factor 12a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| klf12a | dr11_v1_chr1_+_34496855_34496855 | 0.96 | 4.7e-05 | Click! |
Activity profile of klf12a motif
Sorted Z-values of klf12a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_23213320 | 1.04 |
ENSDART00000032996
ENSDART00000137536 |
ppdpfa
|
pancreatic progenitor cell differentiation and proliferation factor a |
| chr13_+_28417297 | 0.97 |
ENSDART00000043658
|
slc2a15a
|
solute carrier family 2 (facilitated glucose transporter), member 15a |
| chr11_-_11301283 | 0.80 |
ENSDART00000113311
ENSDART00000180466 |
col9a1a
|
collagen, type IX, alpha 1a |
| chr1_-_59176949 | 0.70 |
ENSDART00000128742
|
CABZ01118678.1
|
|
| chr10_+_15777064 | 0.69 |
ENSDART00000114483
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr3_+_27027781 | 0.65 |
ENSDART00000065495
|
emp2
|
epithelial membrane protein 2 |
| chr7_+_23907692 | 0.61 |
ENSDART00000045479
|
syt4
|
synaptotagmin IV |
| chr15_+_47161917 | 0.61 |
ENSDART00000167860
|
gap43
|
growth associated protein 43 |
| chr23_-_17470146 | 0.59 |
ENSDART00000002398
|
trim101
|
tripartite motif containing 101 |
| chr4_+_17417111 | 0.59 |
ENSDART00000056005
|
ascl1a
|
achaete-scute family bHLH transcription factor 1a |
| chr25_+_3358701 | 0.57 |
ENSDART00000104877
|
chchd3b
|
coiled-coil-helix-coiled-coil-helix domain containing 3b |
| chr12_+_42436328 | 0.57 |
ENSDART00000167324
|
ebf3a
|
early B cell factor 3a |
| chr2_+_30916188 | 0.56 |
ENSDART00000137012
|
myom1a
|
myomesin 1a (skelemin) |
| chr9_-_296169 | 0.56 |
ENSDART00000165228
|
kif5aa
|
kinesin family member 5A, a |
| chr2_+_59015878 | 0.53 |
ENSDART00000148816
ENSDART00000122795 |
si:ch1073-391i24.1
|
si:ch1073-391i24.1 |
| chr9_+_42095220 | 0.51 |
ENSDART00000148317
ENSDART00000134431 |
pcbp3
|
poly(rC) binding protein 3 |
| chr16_-_17197546 | 0.50 |
ENSDART00000139939
ENSDART00000135146 ENSDART00000063800 ENSDART00000163606 |
gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr10_+_19554604 | 0.49 |
ENSDART00000063806
|
atp6v1b2
|
ATPase H+ transporting V1 subunit B2 |
| chr8_-_4618653 | 0.49 |
ENSDART00000025535
|
sept5a
|
septin 5a |
| chr25_-_25736958 | 0.47 |
ENSDART00000166308
|
cib2
|
calcium and integrin binding family member 2 |
| chr5_+_37056818 | 0.47 |
ENSDART00000036760
|
tppp2
|
tubulin polymerization-promoting protein family member 2 |
| chr25_+_34013093 | 0.47 |
ENSDART00000011967
|
anxa2a
|
annexin A2a |
| chr11_-_25384213 | 0.47 |
ENSDART00000103650
|
mafbb
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Bb |
| chr15_-_19250543 | 0.46 |
ENSDART00000092705
ENSDART00000138895 |
igsf9ba
|
immunoglobulin superfamily, member 9Ba |
| chr20_+_20672163 | 0.46 |
ENSDART00000027758
|
rtn1b
|
reticulon 1b |
| chr7_+_30867008 | 0.45 |
ENSDART00000193106
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr13_+_36764715 | 0.45 |
ENSDART00000111832
ENSDART00000085230 |
atl1
|
atlastin GTPase 1 |
| chr9_+_1654284 | 0.44 |
ENSDART00000062854
|
nfe2l2a
|
nuclear factor, erythroid 2-like 2a |
| chr18_+_44649804 | 0.44 |
ENSDART00000059063
|
ehd2b
|
EH-domain containing 2b |
| chr19_+_48111285 | 0.44 |
ENSDART00000169420
|
nme2b.2
|
NME/NM23 nucleoside diphosphate kinase 2b, tandem duplicate 2 |
| chr2_+_30379650 | 0.43 |
ENSDART00000129542
|
crispld1b
|
cysteine-rich secretory protein LCCL domain containing 1b |
| chr2_-_2020044 | 0.43 |
ENSDART00000024135
|
tubb2
|
tubulin, beta 2A class IIa |
| chr17_+_51764310 | 0.43 |
ENSDART00000157171
|
si:ch211-168d23.3
|
si:ch211-168d23.3 |
| chr15_+_17752928 | 0.42 |
ENSDART00000155314
|
si:ch211-213d14.2
|
si:ch211-213d14.2 |
| chr1_-_51734524 | 0.41 |
ENSDART00000109640
ENSDART00000122628 |
junba
|
JunB proto-oncogene, AP-1 transcription factor subunit a |
| chr4_-_9852318 | 0.41 |
ENSDART00000080702
|
glt8d2
|
glycosyltransferase 8 domain containing 2 |
| chr17_-_4318393 | 0.40 |
ENSDART00000167995
ENSDART00000153824 |
napba
|
N-ethylmaleimide-sensitive factor attachment protein, beta a |
| chr20_-_53366137 | 0.39 |
ENSDART00000146001
|
wasf1
|
WAS protein family, member 1 |
| chr10_-_17988779 | 0.39 |
ENSDART00000132206
ENSDART00000144841 |
si:dkey-242g16.2
|
si:dkey-242g16.2 |
| chr6_-_18976168 | 0.39 |
ENSDART00000170039
|
sept9b
|
septin 9b |
| chr5_+_67812062 | 0.39 |
ENSDART00000158611
|
zgc:175280
|
zgc:175280 |
| chr23_+_10146542 | 0.38 |
ENSDART00000048073
|
zgc:171775
|
zgc:171775 |
| chr6_-_57938043 | 0.38 |
ENSDART00000171073
|
tox2
|
TOX high mobility group box family member 2 |
| chr6_-_17849786 | 0.37 |
ENSDART00000172709
|
rptor
|
regulatory associated protein of MTOR, complex 1 |
| chr10_+_1668106 | 0.37 |
ENSDART00000142278
|
sgsm1b
|
small G protein signaling modulator 1b |
| chr4_-_5597802 | 0.37 |
ENSDART00000136229
|
vegfab
|
vascular endothelial growth factor Ab |
| chr10_+_15777258 | 0.36 |
ENSDART00000140511
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr18_-_3166726 | 0.36 |
ENSDART00000165002
|
aqp11
|
aquaporin 11 |
| chr23_+_44732863 | 0.36 |
ENSDART00000160044
ENSDART00000172268 |
atp1b2a
|
ATPase Na+/K+ transporting subunit beta 2a |
| chr1_-_46663997 | 0.35 |
ENSDART00000134450
|
ebpl
|
emopamil binding protein-like |
| chr19_-_2421793 | 0.35 |
ENSDART00000180238
|
TMEM196 (1 of many)
|
transmembrane protein 196 |
| chr1_-_18803919 | 0.35 |
ENSDART00000020970
|
pgm2
|
phosphoglucomutase 2 |
| chr19_-_42391383 | 0.35 |
ENSDART00000110075
ENSDART00000087002 |
plekho1a
|
pleckstrin homology domain containing, family O member 1a |
| chr13_+_4405282 | 0.34 |
ENSDART00000148280
|
prr18
|
proline rich 18 |
| chr5_-_9824908 | 0.34 |
ENSDART00000169698
|
zgc:158343
|
zgc:158343 |
| chr5_+_38263240 | 0.33 |
ENSDART00000051231
|
gnb2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr16_-_12316979 | 0.33 |
ENSDART00000182392
|
trpv6
|
transient receptor potential cation channel, subfamily V, member 6 |
| chr9_-_1984604 | 0.33 |
ENSDART00000082339
|
hoxd12a
|
homeobox D12a |
| chr21_+_1382078 | 0.33 |
ENSDART00000188463
|
TCF4
|
transcription factor 4 |
| chr19_-_31372896 | 0.32 |
ENSDART00000046609
|
scin
|
scinderin |
| chr3_-_36612877 | 0.32 |
ENSDART00000167164
|
si:dkeyp-72e1.7
|
si:dkeyp-72e1.7 |
| chr1_+_9004719 | 0.31 |
ENSDART00000006211
ENSDART00000137211 |
prkcba
|
protein kinase C, beta a |
| chr5_+_36850650 | 0.31 |
ENSDART00000051186
|
nccrp1
|
non-specific cytotoxic cell receptor protein 1 |
| chr6_+_16031189 | 0.31 |
ENSDART00000015333
|
gbx2
|
gastrulation brain homeobox 2 |
| chr19_+_47290287 | 0.30 |
ENSDART00000078382
|
tpmt.1
|
thiopurine S-methyltransferase, tandem duplicate 1 |
| chr19_-_2420990 | 0.30 |
ENSDART00000181498
|
TMEM196 (1 of many)
|
transmembrane protein 196 |
| chr15_+_24388782 | 0.30 |
ENSDART00000191661
ENSDART00000179995 ENSDART00000111226 |
sez6b
|
seizure related 6 homolog b |
| chr20_+_572037 | 0.29 |
ENSDART00000028062
ENSDART00000152736 ENSDART00000031759 ENSDART00000162198 |
smyd2b
|
SET and MYND domain containing 2b |
| chr21_+_6197223 | 0.29 |
ENSDART00000147716
|
si:dkey-93m18.3
|
si:dkey-93m18.3 |
| chr18_-_46258612 | 0.29 |
ENSDART00000153930
|
si:dkey-244a7.1
|
si:dkey-244a7.1 |
| chr2_+_413370 | 0.29 |
ENSDART00000122138
|
mylk4a
|
myosin light chain kinase family, member 4a |
| chr6_+_59967994 | 0.28 |
ENSDART00000050457
|
zgc:65895
|
zgc:65895 |
| chr7_-_58729894 | 0.27 |
ENSDART00000149347
|
chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr13_-_36391496 | 0.27 |
ENSDART00000100217
ENSDART00000140243 |
actn1
|
actinin, alpha 1 |
| chr17_-_43466317 | 0.26 |
ENSDART00000155313
|
hspa4l
|
heat shock protein 4 like |
| chr6_-_16406210 | 0.26 |
ENSDART00000012023
|
faimb
|
Fas apoptotic inhibitory molecule b |
| chr22_+_10757848 | 0.25 |
ENSDART00000145551
|
tmprss9
|
transmembrane protease, serine 9 |
| chr12_+_47909026 | 0.25 |
ENSDART00000192472
|
tbata
|
thymus, brain and testes associated |
| chr17_+_25332711 | 0.25 |
ENSDART00000082319
|
tmem54a
|
transmembrane protein 54a |
| chr17_+_8925232 | 0.25 |
ENSDART00000036668
|
psmc1a
|
proteasome 26S subunit, ATPase 1a |
| chr18_+_21122818 | 0.25 |
ENSDART00000060015
ENSDART00000060184 |
chka
|
choline kinase alpha |
| chr6_+_16406723 | 0.25 |
ENSDART00000040035
|
ccdc80l1
|
coiled-coil domain containing 80 like 1 |
| chr21_+_30549512 | 0.25 |
ENSDART00000132831
|
rab38c
|
RAB38c, member of RAS oncogene family |
| chr14_+_4151379 | 0.25 |
ENSDART00000160431
|
dhrs13l1
|
dehydrogenase/reductase (SDR family) member 13 like 1 |
| chr8_+_29962635 | 0.25 |
ENSDART00000007640
|
ptch1
|
patched 1 |
| chr24_-_29777413 | 0.25 |
ENSDART00000087724
|
wu:fj05g07
|
wu:fj05g07 |
| chr2_+_23222939 | 0.25 |
ENSDART00000026800
|
kifap3b
|
kinesin-associated protein 3b |
| chr9_+_34425736 | 0.24 |
ENSDART00000135147
|
si:ch211-218d20.15
|
si:ch211-218d20.15 |
| chr6_+_23122789 | 0.24 |
ENSDART00000049226
ENSDART00000067560 |
acox1
|
acyl-CoA oxidase 1, palmitoyl |
| chr14_+_7048930 | 0.23 |
ENSDART00000109138
|
hbegfa
|
heparin-binding EGF-like growth factor a |
| chr10_+_35422802 | 0.23 |
ENSDART00000147303
|
hhla2a.1
|
HERV-H LTR-associating 2a, tandem duplicate 1 |
| chr11_-_42554290 | 0.23 |
ENSDART00000130573
|
atp6ap1la
|
ATPase H+ transporting accessory protein 1 like a |
| chr12_+_13282797 | 0.23 |
ENSDART00000137757
ENSDART00000152397 |
irf9
|
interferon regulatory factor 9 |
| chr13_-_30028103 | 0.23 |
ENSDART00000183889
|
scdb
|
stearoyl-CoA desaturase b |
| chr16_-_42523744 | 0.23 |
ENSDART00000017185
|
tbx20
|
T-box 20 |
| chr3_-_22829710 | 0.23 |
ENSDART00000055659
|
cyb561
|
cytochrome b561 |
| chr25_-_31423493 | 0.23 |
ENSDART00000027661
|
myod1
|
myogenic differentiation 1 |
| chr5_-_46273938 | 0.23 |
ENSDART00000080033
|
si:ch211-130m23.3
|
si:ch211-130m23.3 |
| chr25_-_11088839 | 0.22 |
ENSDART00000154748
|
sv2bb
|
synaptic vesicle glycoprotein 2Bb |
| chr19_+_27479563 | 0.22 |
ENSDART00000049368
ENSDART00000185426 |
atat1
|
alpha tubulin acetyltransferase 1 |
| chr10_+_39952995 | 0.22 |
ENSDART00000183077
|
BX927333.1
|
|
| chr6_-_52156427 | 0.22 |
ENSDART00000082821
|
rims4
|
regulating synaptic membrane exocytosis 4 |
| chr11_+_45287541 | 0.22 |
ENSDART00000165321
ENSDART00000173116 |
pycr1b
|
pyrroline-5-carboxylate reductase 1b |
| chr21_+_11415224 | 0.22 |
ENSDART00000049036
|
zgc:92275
|
zgc:92275 |
| chr22_-_6968756 | 0.21 |
ENSDART00000136636
|
fgfr1bl
|
fibroblast growth factor receptor 1b, like |
| chr10_-_42131408 | 0.21 |
ENSDART00000076693
|
stambpa
|
STAM binding protein a |
| chr2_+_54798689 | 0.21 |
ENSDART00000183426
|
twsg1a
|
twisted gastrulation BMP signaling modulator 1a |
| chr18_-_15467446 | 0.21 |
ENSDART00000187847
|
endouc
|
endonuclease, polyU-specific C |
| chr23_+_31107685 | 0.20 |
ENSDART00000103448
|
tbx18
|
T-box 18 |
| chr8_+_52637507 | 0.20 |
ENSDART00000163830
|
si:dkey-90l8.3
|
si:dkey-90l8.3 |
| chr14_-_24391424 | 0.19 |
ENSDART00000113376
ENSDART00000126894 |
fam13b
|
family with sequence similarity 13, member B |
| chr4_-_11810799 | 0.19 |
ENSDART00000014153
|
cyb5r3
|
cytochrome b5 reductase 3 |
| chr19_+_27479838 | 0.19 |
ENSDART00000103922
|
atat1
|
alpha tubulin acetyltransferase 1 |
| chr19_+_47299212 | 0.19 |
ENSDART00000158262
|
tpmt.1
|
thiopurine S-methyltransferase, tandem duplicate 1 |
| chr25_+_6122823 | 0.19 |
ENSDART00000191824
ENSDART00000067514 |
rbpms2a
|
RNA binding protein with multiple splicing 2a |
| chr14_+_33882973 | 0.19 |
ENSDART00000019396
|
clic2
|
chloride intracellular channel 2 |
| chr18_+_38191346 | 0.19 |
ENSDART00000052703
|
nucb2b
|
nucleobindin 2b |
| chr19_+_8144556 | 0.19 |
ENSDART00000027274
ENSDART00000147218 |
efna3a
|
ephrin-A3a |
| chr19_+_31061718 | 0.19 |
ENSDART00000145971
|
sostdc1b
|
sclerostin domain containing 1b |
| chr18_-_14677936 | 0.19 |
ENSDART00000111995
|
si:dkey-238o13.4
|
si:dkey-238o13.4 |
| chr5_-_33274645 | 0.18 |
ENSDART00000188584
|
kyat1
|
kynurenine aminotransferase 1 |
| chr3_+_36284986 | 0.18 |
ENSDART00000059533
|
wipi1
|
WD repeat domain, phosphoinositide interacting 1 |
| chr22_+_34430310 | 0.18 |
ENSDART00000109860
|
amigo3
|
adhesion molecule with Ig-like domain 3 |
| chr7_+_38762043 | 0.18 |
ENSDART00000036461
|
arhgap1
|
Rho GTPase activating protein 1 |
| chr21_+_30937690 | 0.18 |
ENSDART00000022562
|
rhogb
|
ras homolog family member Gb |
| chr13_+_35339182 | 0.18 |
ENSDART00000019323
|
jag1b
|
jagged 1b |
| chr22_-_881080 | 0.18 |
ENSDART00000185489
|
cept1b
|
choline/ethanolamine phosphotransferase 1b |
| chr23_-_31645760 | 0.18 |
ENSDART00000035031
|
sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr11_-_23458792 | 0.18 |
ENSDART00000032844
|
plekha6
|
pleckstrin homology domain containing, family A member 6 |
| chr3_-_32541033 | 0.17 |
ENSDART00000151476
ENSDART00000055324 |
rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr3_+_14388010 | 0.17 |
ENSDART00000171726
ENSDART00000165452 |
tmem56b
|
transmembrane protein 56b |
| chr20_-_43786515 | 0.16 |
ENSDART00000004601
|
laptm4a
|
lysosomal protein transmembrane 4 alpha |
| chr19_+_22216778 | 0.16 |
ENSDART00000052521
|
nfatc1
|
nuclear factor of activated T cells 1 |
| chr24_-_14712427 | 0.16 |
ENSDART00000176316
|
jph1a
|
junctophilin 1a |
| chr10_+_33382858 | 0.16 |
ENSDART00000063662
|
mdh2
|
malate dehydrogenase 2, NAD (mitochondrial) |
| chr20_-_5254610 | 0.16 |
ENSDART00000057699
|
cyp46a1.2
|
cytochrome P450, family 46, subfamily A, polypeptide 1, tandem duplicate 2 |
| chr11_+_25064519 | 0.16 |
ENSDART00000016181
|
ndrg3a
|
ndrg family member 3a |
| chr16_-_44399335 | 0.16 |
ENSDART00000165058
|
rims2a
|
regulating synaptic membrane exocytosis 2a |
| chr24_-_21923930 | 0.16 |
ENSDART00000131944
|
tagln3b
|
transgelin 3b |
| chr2_+_25657958 | 0.16 |
ENSDART00000161407
|
tnika
|
TRAF2 and NCK interacting kinase a |
| chr2_+_25560556 | 0.16 |
ENSDART00000133623
|
pld1a
|
phospholipase D1a |
| chr6_+_34512313 | 0.16 |
ENSDART00000102554
|
lrp8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr25_+_7492663 | 0.16 |
ENSDART00000166496
|
cat
|
catalase |
| chr4_-_9557186 | 0.16 |
ENSDART00000150569
|
shank3b
|
SH3 and multiple ankyrin repeat domains 3b |
| chr17_+_53250802 | 0.16 |
ENSDART00000143819
|
VASH1
|
vasohibin 1 |
| chr6_+_34511886 | 0.16 |
ENSDART00000179450
|
lrp8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr3_-_47876427 | 0.15 |
ENSDART00000180844
ENSDART00000124480 |
adgrl1a
|
adhesion G protein-coupled receptor L1a |
| chr14_+_33264303 | 0.15 |
ENSDART00000130680
ENSDART00000075187 |
pdzd11
|
PDZ domain containing 11 |
| chr7_-_6592142 | 0.15 |
ENSDART00000160137
|
kcnj10a
|
potassium inwardly-rectifying channel, subfamily J, member 10a |
| chr20_-_46554440 | 0.15 |
ENSDART00000043298
ENSDART00000060680 |
fosab
|
v-fos FBJ murine osteosarcoma viral oncogene homolog Ab |
| chr21_+_25236297 | 0.15 |
ENSDART00000112783
|
tmem45b
|
transmembrane protein 45B |
| chr4_-_11810365 | 0.15 |
ENSDART00000150529
|
cyb5r3
|
cytochrome b5 reductase 3 |
| chr22_-_38224315 | 0.14 |
ENSDART00000165430
ENSDART00000140968 |
hps3
|
Hermansky-Pudlak syndrome 3 |
| chr5_-_2689753 | 0.14 |
ENSDART00000172699
|
gng10
|
guanine nucleotide binding protein (G protein), gamma 10 |
| chr12_+_48815988 | 0.14 |
ENSDART00000149089
|
anxa11b
|
annexin A11b |
| chr22_-_29586608 | 0.14 |
ENSDART00000059869
|
adra2a
|
adrenoceptor alpha 2A |
| chr23_-_32129569 | 0.14 |
ENSDART00000167761
ENSDART00000139569 |
zgc:92658
|
zgc:92658 |
| chr19_+_342094 | 0.14 |
ENSDART00000151013
ENSDART00000187622 |
ensaa
|
endosulfine alpha a |
| chr4_+_3980247 | 0.14 |
ENSDART00000049194
|
gpr37b
|
G protein-coupled receptor 37b |
| chr23_-_17429775 | 0.14 |
ENSDART00000043076
|
ppdpfb
|
pancreatic progenitor cell differentiation and proliferation factor b |
| chr14_-_2602445 | 0.14 |
ENSDART00000166910
|
etf1a
|
eukaryotic translation termination factor 1a |
| chr5_-_50992690 | 0.14 |
ENSDART00000149553
ENSDART00000097460 ENSDART00000192021 |
hmgcra
|
3-hydroxy-3-methylglutaryl-CoA reductase a |
| chr24_-_36238054 | 0.14 |
ENSDART00000155725
|
tmem241
|
transmembrane protein 241 |
| chr2_+_25658112 | 0.14 |
ENSDART00000051234
|
tnika
|
TRAF2 and NCK interacting kinase a |
| chr25_+_19008497 | 0.14 |
ENSDART00000104420
|
samm50
|
SAMM50 sorting and assembly machinery component |
| chr3_+_22578369 | 0.13 |
ENSDART00000187695
ENSDART00000182678 ENSDART00000112270 |
tanc2a
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2a |
| chr1_-_8653385 | 0.13 |
ENSDART00000193041
|
actb1
|
actin, beta 1 |
| chr25_+_3788443 | 0.13 |
ENSDART00000189747
|
chid1
|
chitinase domain containing 1 |
| chr13_+_16279890 | 0.13 |
ENSDART00000101775
ENSDART00000057948 |
anxa11a
|
annexin A11a |
| chr3_+_13511984 | 0.13 |
ENSDART00000158942
|
CABZ01015475.1
|
|
| chr11_+_35364445 | 0.13 |
ENSDART00000125766
|
camkvb
|
CaM kinase-like vesicle-associated b |
| chr5_-_72136548 | 0.13 |
ENSDART00000007827
|
spra
|
sepiapterin reductase a |
| chr20_-_54381034 | 0.13 |
ENSDART00000136779
|
entpd5b
|
ectonucleoside triphosphate diphosphohydrolase 5b |
| chr20_-_9580446 | 0.13 |
ENSDART00000014168
|
zfp36l1b
|
zinc finger protein 36, C3H type-like 1b |
| chr3_+_5575313 | 0.13 |
ENSDART00000134693
ENSDART00000101807 |
si:ch211-106h11.3
|
si:ch211-106h11.3 |
| chr7_-_18601206 | 0.13 |
ENSDART00000111636
|
DTX4
|
si:ch211-119e14.2 |
| chr3_+_19299309 | 0.12 |
ENSDART00000046297
ENSDART00000146955 |
ldlra
|
low density lipoprotein receptor a |
| chr22_+_661711 | 0.12 |
ENSDART00000113795
|
elf3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr12_+_6391243 | 0.12 |
ENSDART00000152765
|
prkg1b
|
protein kinase, cGMP-dependent, type Ib |
| chr14_+_46410766 | 0.12 |
ENSDART00000032342
|
anxa5a
|
annexin A5a |
| chr25_-_6049339 | 0.12 |
ENSDART00000075184
|
snx1a
|
sorting nexin 1a |
| chr14_+_12109535 | 0.12 |
ENSDART00000054620
ENSDART00000121913 |
kctd12b
|
potassium channel tetramerisation domain containing 12b |
| chr8_-_1956632 | 0.12 |
ENSDART00000131602
|
si:dkey-178e17.1
|
si:dkey-178e17.1 |
| chr10_+_1638876 | 0.12 |
ENSDART00000184484
ENSDART00000060946 ENSDART00000181251 |
sgsm1b
|
small G protein signaling modulator 1b |
| chr8_-_17167819 | 0.12 |
ENSDART00000135042
ENSDART00000143920 |
mrps36
|
mitochondrial ribosomal protein S36 |
| chr25_-_1720736 | 0.11 |
ENSDART00000097256
|
SLC6A13
|
solute carrier family 6 member 13 |
| chr13_+_421231 | 0.11 |
ENSDART00000188212
ENSDART00000017854 |
lgi1a
|
leucine-rich, glioma inactivated 1a |
| chr21_-_32060993 | 0.11 |
ENSDART00000131651
|
si:ch211-160j14.2
|
si:ch211-160j14.2 |
| chr25_+_25737386 | 0.11 |
ENSDART00000108476
|
lrrc61
|
leucine rich repeat containing 61 |
| chr3_+_472158 | 0.11 |
ENSDART00000134971
|
zgc:194659
|
zgc:194659 |
| chr10_+_4875262 | 0.11 |
ENSDART00000165942
|
palm2
|
paralemmin 2 |
| chr1_+_42874410 | 0.11 |
ENSDART00000153506
|
ctnna2
|
catenin (cadherin-associated protein), alpha 2 |
| chr24_+_41931585 | 0.11 |
ENSDART00000130310
|
epb41l3a
|
erythrocyte membrane protein band 4.1-like 3a |
| chr17_-_31695217 | 0.11 |
ENSDART00000104332
ENSDART00000143090 |
lin52
|
lin-52 DREAM MuvB core complex component |
| chr14_+_12110020 | 0.11 |
ENSDART00000192462
|
kctd12b
|
potassium channel tetramerisation domain containing 12b |
| chr13_-_36621926 | 0.11 |
ENSDART00000057155
|
cdkn3
|
cyclin-dependent kinase inhibitor 3 |
| chr5_-_68022631 | 0.10 |
ENSDART00000143199
|
wasf3a
|
WAS protein family, member 3a |
| chr11_+_29790626 | 0.10 |
ENSDART00000067822
|
dynlt3
|
dynein, light chain, Tctex-type 3 |
| chr25_+_4635355 | 0.10 |
ENSDART00000021120
|
pgghg
|
protein-glucosylgalactosylhydroxylysine glucosidase |
| chr13_+_49175947 | 0.10 |
ENSDART00000056927
|
egln1a
|
egl-9 family hypoxia-inducible factor 1a |
Network of associatons between targets according to the STRING database.
First level regulatory network of klf12a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0001977 | renal system process involved in regulation of blood volume(GO:0001977) renal system process involved in regulation of systemic arterial blood pressure(GO:0003071) regulation of glomerular filtration(GO:0003093) |
| 0.1 | 0.6 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.1 | 0.6 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.1 | 0.5 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.1 | 0.4 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.1 | 0.3 | GO:0045023 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.1 | 0.2 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.1 | 0.4 | GO:1902023 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.1 | 0.2 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.1 | 0.2 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.1 | 0.2 | GO:0060043 | regulation of cardiac muscle tissue growth(GO:0055021) regulation of cardiac muscle cell proliferation(GO:0060043) |
| 0.1 | 0.3 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.1 | 0.2 | GO:0007008 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.2 | GO:0032534 | regulation of microvillus organization(GO:0032530) regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.4 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.0 | 0.6 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.0 | 0.2 | GO:0051148 | smooth muscle cell differentiation(GO:0051145) negative regulation of muscle cell differentiation(GO:0051148) |
| 0.0 | 0.3 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.4 | GO:0060753 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.5 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.2 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.3 | GO:1904861 | excitatory synapse assembly(GO:1904861) |
| 0.0 | 0.1 | GO:0006843 | mitochondrial citrate transport(GO:0006843) |
| 0.0 | 0.1 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.0 | 0.3 | GO:0021871 | forebrain regionalization(GO:0021871) |
| 0.0 | 0.1 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.0 | 0.1 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.0 | 0.4 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.2 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.0 | 0.2 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.3 | GO:1902041 | regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902041) negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.0 | 0.6 | GO:0007007 | inner mitochondrial membrane organization(GO:0007007) |
| 0.0 | 0.0 | GO:0032770 | regulation of monooxygenase activity(GO:0032768) positive regulation of monooxygenase activity(GO:0032770) |
| 0.0 | 0.1 | GO:0036268 | swimming(GO:0036268) |
| 0.0 | 0.2 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.1 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.0 | 0.1 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 1.2 | GO:0031017 | exocrine pancreas development(GO:0031017) exocrine system development(GO:0035272) |
| 0.0 | 0.1 | GO:0015919 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.4 | GO:0010962 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.0 | 0.2 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.4 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.6 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.4 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.2 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.1 | GO:0070292 | N-acylphosphatidylethanolamine metabolic process(GO:0070292) |
| 0.0 | 0.3 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.2 | GO:0046337 | phosphatidylethanolamine biosynthetic process(GO:0006646) phosphatidylethanolamine metabolic process(GO:0046337) |
| 0.0 | 0.2 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.2 | GO:0035306 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.0 | 0.5 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 0.2 | GO:0070189 | kynurenine metabolic process(GO:0070189) |
| 0.0 | 0.2 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 0.3 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.5 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.0 | 0.2 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.1 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.0 | 0.2 | GO:0015936 | coenzyme A metabolic process(GO:0015936) |
| 0.0 | 0.0 | GO:1990868 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) negative regulation of lymphocyte migration(GO:2000402) negative regulation of T cell migration(GO:2000405) |
| 0.0 | 0.2 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.0 | 0.1 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.0 | 0.2 | GO:0042542 | response to hydrogen peroxide(GO:0042542) |
| 0.0 | 0.2 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.7 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 0.3 | GO:0051014 | actin filament severing(GO:0051014) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 0.4 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 0.6 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.4 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.7 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 0.4 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.5 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.1 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.0 | 0.6 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.3 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.9 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.2 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.0 | 0.3 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.1 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.4 | GO:0099569 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic cytoskeleton(GO:0099569) |
| 0.0 | 0.4 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 1.4 | GO:0044309 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 0.2 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.4 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.1 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 1.0 | GO:0005871 | kinesin complex(GO:0005871) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0008119 | thiopurine S-methyltransferase activity(GO:0008119) |
| 0.1 | 0.5 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 0.4 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 0.3 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.1 | 0.2 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.1 | 0.4 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 1.5 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 0.2 | GO:0047804 | cysteine-S-conjugate beta-lyase activity(GO:0047804) |
| 0.1 | 0.2 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.1 | 0.8 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 0.5 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.4 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.1 | 0.4 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 0.2 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.3 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.2 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.3 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.5 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.0 | 0.2 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.2 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.2 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.1 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.0 | 0.1 | GO:0000035 | acyl binding(GO:0000035) |
| 0.0 | 0.3 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.2 | GO:0033781 | cholesterol 24-hydroxylase activity(GO:0033781) |
| 0.0 | 0.4 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.1 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.2 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.2 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.3 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.3 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.2 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.1 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.0 | 0.4 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.2 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.1 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.6 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.3 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.2 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0016433 | rRNA (adenine) methyltransferase activity(GO:0016433) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.2 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.3 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.4 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.6 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.7 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.3 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |