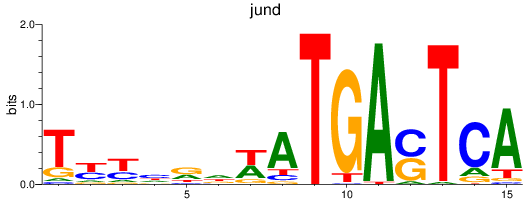
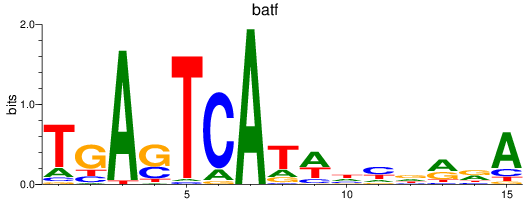
Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
Results for jund_batf
Z-value: 1.04
Transcription factors associated with jund_batf
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
jund
|
ENSDARG00000067850 | JunD proto-oncogene, AP-1 transcription factor subunit |
|
batf
|
ENSDARG00000011818 | basic leucine zipper transcription factor, ATF-like |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| batf | dr11_v1_chr20_+_46572550_46572550 | -0.82 | 6.9e-03 | Click! |
| jund | dr11_v1_chr2_-_56131312_56131312 | 0.14 | 7.2e-01 | Click! |
Activity profile of jund_batf motif
Sorted Z-values of jund_batf motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_-_26595027 | 1.96 |
ENSDART00000184162
|
CABZ01072309.1
|
|
| chr3_+_31621774 | 1.94 |
ENSDART00000076636
|
fzd2
|
frizzled class receptor 2 |
| chr3_-_26017592 | 1.59 |
ENSDART00000030890
|
hmox1a
|
heme oxygenase 1a |
| chr22_+_16497670 | 1.58 |
ENSDART00000014330
|
ier5
|
immediate early response 5 |
| chr3_-_26017831 | 1.40 |
ENSDART00000179982
|
hmox1a
|
heme oxygenase 1a |
| chr3_+_24197934 | 1.39 |
ENSDART00000055609
|
atf4b
|
activating transcription factor 4b |
| chr1_-_56176976 | 1.27 |
ENSDART00000052688
|
c3a.1
|
complement component c3a, duplicate 1 |
| chr20_+_572037 | 1.25 |
ENSDART00000028062
ENSDART00000152736 ENSDART00000031759 ENSDART00000162198 |
smyd2b
|
SET and MYND domain containing 2b |
| chr6_+_49095646 | 1.25 |
ENSDART00000103385
|
slc25a55a
|
solute carrier family 25, member 55a |
| chr7_-_26436436 | 1.24 |
ENSDART00000019035
ENSDART00000123395 |
her8a
|
hairy-related 8a |
| chr12_+_9551667 | 1.16 |
ENSDART00000048493
ENSDART00000166178 |
p4ha1a
|
prolyl 4-hydroxylase, alpha polypeptide I a |
| chr21_+_22845317 | 1.08 |
ENSDART00000065555
|
birc2
|
baculoviral IAP repeat containing 2 |
| chr1_-_22834824 | 1.05 |
ENSDART00000043556
|
ldb2b
|
LIM domain binding 2b |
| chr7_-_52963493 | 1.04 |
ENSDART00000052029
|
cart3
|
cocaine- and amphetamine-regulated transcript 3 |
| chr7_-_24236364 | 0.94 |
ENSDART00000010124
|
slc7a8a
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8a |
| chr21_+_5888641 | 0.89 |
ENSDART00000091331
|
prodha
|
proline dehydrogenase (oxidase) 1a |
| chr21_-_44512893 | 0.88 |
ENSDART00000166853
|
zgc:136410
|
zgc:136410 |
| chr21_+_17024002 | 0.88 |
ENSDART00000080628
|
arpc3
|
actin related protein 2/3 complex, subunit 3 |
| chr8_+_39634114 | 0.83 |
ENSDART00000144293
|
msi1
|
musashi RNA-binding protein 1 |
| chr19_-_21716593 | 0.82 |
ENSDART00000155126
|
znf516
|
zinc finger protein 516 |
| chr23_-_1017428 | 0.82 |
ENSDART00000110588
ENSDART00000183158 |
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr16_+_42830152 | 0.75 |
ENSDART00000159730
|
polr3glb
|
polymerase (RNA) III (DNA directed) polypeptide G like b |
| chr5_-_30924347 | 0.74 |
ENSDART00000111749
ENSDART00000086564 ENSDART00000153909 |
spns2
|
spinster homolog 2 (Drosophila) |
| chr13_+_24280380 | 0.74 |
ENSDART00000184115
|
acta1b
|
actin, alpha 1b, skeletal muscle |
| chr23_+_20863145 | 0.73 |
ENSDART00000129992
|
pax7b
|
paired box 7b |
| chr11_-_39202915 | 0.73 |
ENSDART00000105133
|
wnt4a
|
wingless-type MMTV integration site family, member 4a |
| chr15_-_21877726 | 0.72 |
ENSDART00000127819
ENSDART00000145646 ENSDART00000100897 ENSDART00000144739 |
zgc:162608
|
zgc:162608 |
| chr18_-_46010 | 0.70 |
ENSDART00000052641
|
gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr23_+_9067131 | 0.69 |
ENSDART00000144533
|
ccm2l
|
cerebral cavernous malformation 2-like |
| chr3_-_39488482 | 0.66 |
ENSDART00000135192
|
zgc:100868
|
zgc:100868 |
| chr3_-_16289826 | 0.61 |
ENSDART00000131972
|
bckdhbl
|
branched chain keto acid dehydrogenase E1, beta polypeptide, like |
| chr20_-_42972599 | 0.60 |
ENSDART00000100751
|
pomcb
|
proopiomelanocortin b |
| chr1_+_135903 | 0.60 |
ENSDART00000124837
|
f10
|
coagulation factor X |
| chr13_+_1872767 | 0.59 |
ENSDART00000161162
|
bmp5
|
bone morphogenetic protein 5 |
| chr5_+_38276582 | 0.59 |
ENSDART00000158532
|
gnb2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr6_+_598669 | 0.58 |
ENSDART00000151009
|
si:ch73-379f7.4
|
si:ch73-379f7.4 |
| chr3_-_39488639 | 0.57 |
ENSDART00000161644
|
zgc:100868
|
zgc:100868 |
| chr8_+_25900049 | 0.57 |
ENSDART00000124300
ENSDART00000127618 ENSDART00000024009 |
rhoab
|
ras homolog gene family, member Ab |
| chr16_-_38518270 | 0.56 |
ENSDART00000131899
|
rspo2
|
R-spondin 2 |
| chr24_+_35827766 | 0.56 |
ENSDART00000144700
|
si:dkeyp-7a3.1
|
si:dkeyp-7a3.1 |
| chr8_+_8973425 | 0.54 |
ENSDART00000066107
|
bcap31
|
B cell receptor associated protein 31 |
| chr6_-_31224563 | 0.53 |
ENSDART00000104616
|
lepr
|
leptin receptor |
| chr21_+_25765734 | 0.52 |
ENSDART00000021664
|
cldnb
|
claudin b |
| chr24_+_13277573 | 0.52 |
ENSDART00000137886
|
si:ch211-171b20.3
|
si:ch211-171b20.3 |
| chr9_+_2499627 | 0.52 |
ENSDART00000160782
|
wipf1a
|
WAS/WASL interacting protein family, member 1a |
| chr4_+_68087932 | 0.52 |
ENSDART00000150494
|
znf1096
|
zinc finger protein 1096 |
| chr4_+_1530287 | 0.51 |
ENSDART00000067446
|
slc38a4
|
solute carrier family 38, member 4 |
| chr22_+_38173960 | 0.50 |
ENSDART00000010537
|
cp
|
ceruloplasmin |
| chr25_-_18330503 | 0.50 |
ENSDART00000104496
|
dusp6
|
dual specificity phosphatase 6 |
| chr18_+_24921587 | 0.49 |
ENSDART00000191345
|
rgma
|
repulsive guidance molecule family member a |
| chr6_+_56141852 | 0.49 |
ENSDART00000149665
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr20_+_25340814 | 0.48 |
ENSDART00000063028
|
ctgfa
|
connective tissue growth factor a |
| chr23_-_1017605 | 0.47 |
ENSDART00000138290
|
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr5_-_41531629 | 0.45 |
ENSDART00000051082
|
akr1a1a
|
aldo-keto reductase family 1, member A1a (aldehyde reductase) |
| chr4_+_68088529 | 0.43 |
ENSDART00000159155
|
znf1096
|
zinc finger protein 1096 |
| chr6_+_7421898 | 0.42 |
ENSDART00000043946
|
ccdc65
|
coiled-coil domain containing 65 |
| chr13_+_35637875 | 0.41 |
ENSDART00000180657
|
thbs2a
|
thrombospondin 2a |
| chr1_+_54835131 | 0.41 |
ENSDART00000145070
|
si:ch211-196h16.4
|
si:ch211-196h16.4 |
| chr24_+_31361407 | 0.39 |
ENSDART00000162668
|
cremb
|
cAMP responsive element modulator b |
| chr14_-_976912 | 0.39 |
ENSDART00000114053
|
arsia
|
arylsulfatase family, member Ia |
| chr3_-_16055432 | 0.38 |
ENSDART00000123621
ENSDART00000023859 |
atp6v0ca
|
ATPase H+ transporting V0 subunit ca |
| chr13_+_35339182 | 0.36 |
ENSDART00000019323
|
jag1b
|
jagged 1b |
| chr8_-_40276035 | 0.35 |
ENSDART00000185301
ENSDART00000187610 ENSDART00000053158 ENSDART00000162020 |
kdm2ba
|
lysine (K)-specific demethylase 2Ba |
| chr18_+_45114392 | 0.34 |
ENSDART00000172328
|
large2
|
LARGE xylosyl- and glucuronyltransferase 2 |
| chr16_-_32184460 | 0.33 |
ENSDART00000102027
|
kpna5
|
karyopherin alpha 5 (importin alpha 6) |
| chr16_-_50203058 | 0.33 |
ENSDART00000154570
|
vsig10l
|
V-set and immunoglobulin domain containing 10 like |
| chr17_-_52091999 | 0.33 |
ENSDART00000019766
|
tgfb3
|
transforming growth factor, beta 3 |
| chr14_+_49382180 | 0.33 |
ENSDART00000158329
|
tnip1
|
TNFAIP3 interacting protein 1 |
| chr10_+_31953502 | 0.33 |
ENSDART00000185634
|
lhfpl6
|
LHFPL tetraspan subfamily member 6 |
| chr9_-_1978090 | 0.32 |
ENSDART00000082344
|
hoxd11a
|
homeobox D11a |
| chr10_-_43928937 | 0.32 |
ENSDART00000168014
ENSDART00000086220 |
mctp1b
|
multiple C2 domains, transmembrane 1b |
| chr3_+_31177972 | 0.31 |
ENSDART00000185954
|
clec19a
|
C-type lectin domain containing 19A |
| chr23_+_39558508 | 0.31 |
ENSDART00000017902
|
camk1gb
|
calcium/calmodulin-dependent protein kinase IGb |
| chr22_+_5851122 | 0.30 |
ENSDART00000082044
|
zmp:0000001161
|
zmp:0000001161 |
| chr8_+_27807266 | 0.30 |
ENSDART00000170037
|
capza1b
|
capping protein (actin filament) muscle Z-line, alpha 1b |
| chr3_+_37707432 | 0.30 |
ENSDART00000151236
|
map3k14a
|
mitogen-activated protein kinase kinase kinase 14a |
| chr8_-_52745141 | 0.29 |
ENSDART00000168359
ENSDART00000168252 |
fgf17
|
fibroblast growth factor 17 |
| chr20_-_34127415 | 0.29 |
ENSDART00000010028
|
ptgs2b
|
prostaglandin-endoperoxide synthase 2b |
| chr3_+_24986145 | 0.29 |
ENSDART00000055428
|
cbx7a
|
chromobox homolog 7a |
| chr17_-_33714636 | 0.29 |
ENSDART00000188500
|
DNAL1
|
si:dkey-84k17.3 |
| chr7_-_35409027 | 0.29 |
ENSDART00000128334
|
lpcat2
|
lysophosphatidylcholine acyltransferase 2 |
| chr13_-_21701323 | 0.29 |
ENSDART00000164112
|
si:dkey-191g9.7
|
si:dkey-191g9.7 |
| chr7_-_35408618 | 0.28 |
ENSDART00000074963
|
lpcat2
|
lysophosphatidylcholine acyltransferase 2 |
| chr21_+_8341774 | 0.28 |
ENSDART00000129749
ENSDART00000055325 ENSDART00000133804 |
psmb7
|
proteasome subunit beta 7 |
| chr24_-_21404367 | 0.28 |
ENSDART00000152093
|
atp8a2
|
ATPase phospholipid transporting 8A2 |
| chr23_+_30967686 | 0.28 |
ENSDART00000144485
|
si:ch211-197l9.2
|
si:ch211-197l9.2 |
| chr4_-_45146463 | 0.28 |
ENSDART00000187483
|
znf1046
|
zinc finger protein 1046 |
| chr5_+_54585431 | 0.27 |
ENSDART00000171225
|
npr2
|
natriuretic peptide receptor 2 |
| chr1_-_40086978 | 0.27 |
ENSDART00000136990
|
si:dkey-117m1.4
|
si:dkey-117m1.4 |
| chr18_-_28938912 | 0.26 |
ENSDART00000136201
|
si:ch211-174j14.2
|
si:ch211-174j14.2 |
| chr4_+_28356606 | 0.26 |
ENSDART00000192995
|
si:ch73-263o4.4
|
si:ch73-263o4.4 |
| chr1_-_35694978 | 0.26 |
ENSDART00000136157
|
si:dkey-27h10.2
|
si:dkey-27h10.2 |
| chr13_-_50624743 | 0.26 |
ENSDART00000167949
|
vox
|
ventral homeobox |
| chr10_+_31248036 | 0.25 |
ENSDART00000193574
|
robo4
|
roundabout, axon guidance receptor, homolog 4 (Drosophila) |
| chr3_-_61185746 | 0.25 |
ENSDART00000028219
|
pvalb4
|
parvalbumin 4 |
| chr5_-_30487822 | 0.25 |
ENSDART00000189288
ENSDART00000183201 |
phldb1a
|
pleckstrin homology-like domain, family B, member 1a |
| chr15_-_20933574 | 0.25 |
ENSDART00000152648
ENSDART00000152448 ENSDART00000152244 |
usp2a
|
ubiquitin specific peptidase 2a |
| chr18_+_9362455 | 0.25 |
ENSDART00000187025
|
sema3ab
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Ab |
| chr14_+_48062180 | 0.25 |
ENSDART00000056713
|
ppid
|
peptidylprolyl isomerase D |
| chr9_-_24244383 | 0.25 |
ENSDART00000182407
|
cavin2a
|
caveolae associated protein 2a |
| chr13_-_42749916 | 0.24 |
ENSDART00000140019
|
capn2a
|
calpain 2, (m/II) large subunit a |
| chr20_+_34868933 | 0.24 |
ENSDART00000153006
|
ankef1a
|
ankyrin repeat and EF-hand domain containing 1a |
| chr9_+_38372216 | 0.24 |
ENSDART00000141895
|
plcd4b
|
phospholipase C, delta 4b |
| chr19_-_33212023 | 0.24 |
ENSDART00000189209
|
trib1
|
tribbles pseudokinase 1 |
| chr4_-_30057608 | 0.23 |
ENSDART00000184429
|
si:rp71-7l19.2
|
si:rp71-7l19.2 |
| chr25_-_15496485 | 0.22 |
ENSDART00000140245
|
si:dkeyp-67e1.6
|
si:dkeyp-67e1.6 |
| chr5_-_43959972 | 0.22 |
ENSDART00000180517
|
si:ch211-204c21.1
|
si:ch211-204c21.1 |
| chr13_-_43108693 | 0.22 |
ENSDART00000164439
|
si:ch211-106f21.1
|
si:ch211-106f21.1 |
| chr14_-_17588345 | 0.21 |
ENSDART00000143486
|
selenot2
|
selenoprotein T, 2 |
| chr17_-_23727978 | 0.21 |
ENSDART00000079600
|
minpp1a
|
multiple inositol-polyphosphate phosphatase 1a |
| chr19_+_48102560 | 0.21 |
ENSDART00000164464
|
utp18
|
UTP18 small subunit (SSU) processome component |
| chr13_-_20381485 | 0.20 |
ENSDART00000131351
|
si:ch211-270n8.1
|
si:ch211-270n8.1 |
| chr5_+_17624463 | 0.20 |
ENSDART00000183869
ENSDART00000081064 |
fbrsl1
|
fibrosin-like 1 |
| chr25_+_27873836 | 0.20 |
ENSDART00000163801
|
iqub
|
IQ motif and ubiquitin domain containing |
| chr2_+_4075073 | 0.20 |
ENSDART00000165634
ENSDART00000158602 ENSDART00000163507 |
mib1
|
mindbomb E3 ubiquitin protein ligase 1 |
| chr3_-_16493528 | 0.20 |
ENSDART00000142099
|
si:ch211-23l10.3
|
si:ch211-23l10.3 |
| chr6_+_41957280 | 0.20 |
ENSDART00000050114
|
oxtr
|
oxytocin receptor |
| chr6_-_43449013 | 0.19 |
ENSDART00000122423
|
eevs
|
2-epi-5-epi-valiolone synthase |
| chr7_+_25036188 | 0.19 |
ENSDART00000163957
ENSDART00000169749 |
sb:cb1058
|
sb:cb1058 |
| chr22_-_29336268 | 0.19 |
ENSDART00000132776
ENSDART00000186351 ENSDART00000121599 |
pdgfba
|
platelet-derived growth factor beta polypeptide a |
| chr8_+_27807974 | 0.19 |
ENSDART00000078509
|
capza1b
|
capping protein (actin filament) muscle Z-line, alpha 1b |
| chr14_-_32884138 | 0.18 |
ENSDART00000105726
|
slc25a5
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 |
| chr9_-_23181062 | 0.18 |
ENSDART00000192578
|
lypd6b
|
LY6/PLAUR domain containing 6B |
| chr21_+_9628854 | 0.18 |
ENSDART00000161753
ENSDART00000160711 |
mapk10
|
mitogen-activated protein kinase 10 |
| chr11_-_40101246 | 0.18 |
ENSDART00000161083
|
tnfrsf9b
|
tumor necrosis factor receptor superfamily, member 9b |
| chr1_+_7517454 | 0.18 |
ENSDART00000016139
|
lancl1
|
LanC antibiotic synthetase component C-like 1 (bacterial) |
| chr15_+_29123031 | 0.18 |
ENSDART00000133988
ENSDART00000060030 |
zgc:101731
|
zgc:101731 |
| chr23_+_24124684 | 0.17 |
ENSDART00000144478
|
si:dkey-21o19.2
|
si:dkey-21o19.2 |
| chr18_+_5213338 | 0.16 |
ENSDART00000033574
|
slc24a5
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 5 |
| chr8_+_43053519 | 0.16 |
ENSDART00000147178
|
prnpa
|
prion protein a |
| chr1_+_14253118 | 0.16 |
ENSDART00000161996
|
cxcl8a
|
chemokine (C-X-C motif) ligand 8a |
| chr23_-_44848961 | 0.16 |
ENSDART00000136839
|
wu:fb72h05
|
wu:fb72h05 |
| chr25_+_27873671 | 0.16 |
ENSDART00000088817
|
iqub
|
IQ motif and ubiquitin domain containing |
| chr4_-_69189894 | 0.15 |
ENSDART00000169596
|
si:ch211-209j12.1
|
si:ch211-209j12.1 |
| chr4_-_76270779 | 0.15 |
ENSDART00000183709
ENSDART00000192689 |
si:ch211-106j21.4
|
si:ch211-106j21.4 |
| chr10_+_42733210 | 0.15 |
ENSDART00000189832
|
CABZ01063556.1
|
|
| chr4_+_33654247 | 0.15 |
ENSDART00000192537
|
si:dkey-84h14.1
|
si:dkey-84h14.1 |
| chr22_-_12746539 | 0.15 |
ENSDART00000175374
|
plcd4a
|
phospholipase C, delta 4a |
| chr13_-_50624173 | 0.15 |
ENSDART00000184181
|
vox
|
ventral homeobox |
| chr12_+_28117365 | 0.15 |
ENSDART00000066290
|
UTS2R
|
urotensin 2 receptor |
| chr20_-_29418620 | 0.15 |
ENSDART00000172634
|
ryr3
|
ryanodine receptor 3 |
| chr10_+_40700311 | 0.15 |
ENSDART00000157650
ENSDART00000138342 |
taar19n
|
trace amine associated receptor 19n |
| chr19_-_5669122 | 0.14 |
ENSDART00000112211
|
si:ch211-264f5.2
|
si:ch211-264f5.2 |
| chr18_-_5692292 | 0.14 |
ENSDART00000121503
|
cplx3b
|
complexin 3b |
| chr25_+_10410620 | 0.14 |
ENSDART00000151886
|
ehf
|
ets homologous factor |
| chr4_+_76957765 | 0.13 |
ENSDART00000129607
|
si:dkey-240n22.6
|
si:dkey-240n22.6 |
| chr7_-_37563883 | 0.13 |
ENSDART00000148805
|
nod2
|
nucleotide-binding oligomerization domain containing 2 |
| chr4_-_76091052 | 0.13 |
ENSDART00000124979
|
si:ch211-232d10.1
|
si:ch211-232d10.1 |
| chr17_+_23770848 | 0.12 |
ENSDART00000079646
|
kcnk18
|
potassium channel, subfamily K, member 18 |
| chr17_+_18117358 | 0.12 |
ENSDART00000144894
|
bcl11ba
|
B cell CLL/lymphoma 11Ba |
| chr2_-_19109304 | 0.12 |
ENSDART00000168028
|
si:dkey-225f23.5
|
si:dkey-225f23.5 |
| chr12_+_7497882 | 0.12 |
ENSDART00000134608
|
phyhiplb
|
phytanoyl-CoA 2-hydroxylase interacting protein-like b |
| chr23_-_10914275 | 0.11 |
ENSDART00000112965
|
pdzrn3a
|
PDZ domain containing RING finger 3a |
| chr14_+_10461264 | 0.11 |
ENSDART00000081095
|
cysltr1
|
cysteinyl leukotriene receptor 1 |
| chr25_+_25123385 | 0.11 |
ENSDART00000163892
|
ldha
|
lactate dehydrogenase A4 |
| chr13_+_8987957 | 0.11 |
ENSDART00000148144
|
hcar1-3
|
hydroxycarboxylic acid receptor 1-3 |
| chr4_+_54899568 | 0.11 |
ENSDART00000162786
|
si:dkey-56m15.8
|
si:dkey-56m15.8 |
| chr12_+_1455147 | 0.11 |
ENSDART00000018752
|
cops3
|
COP9 signalosome subunit 3 |
| chr20_+_20726231 | 0.11 |
ENSDART00000147112
|
zgc:193541
|
zgc:193541 |
| chr4_+_71014655 | 0.09 |
ENSDART00000170837
|
si:dkeyp-80d11.14
|
si:dkeyp-80d11.14 |
| chr4_+_47436126 | 0.09 |
ENSDART00000157555
|
si:dkey-124l13.1
|
si:dkey-124l13.1 |
| chr20_-_35040041 | 0.09 |
ENSDART00000131919
|
kif26bb
|
kinesin family member 26Bb |
| chr4_-_36476889 | 0.09 |
ENSDART00000163956
|
si:ch211-263l8.1
|
si:ch211-263l8.1 |
| chr3_-_4760384 | 0.09 |
ENSDART00000108810
|
CABZ01046997.1
|
|
| chr2_-_16224083 | 0.08 |
ENSDART00000165953
|
arhgef4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr8_+_25034544 | 0.08 |
ENSDART00000123300
|
ngrn
|
neugrin, neurite outgrowth associated |
| chr7_-_13906409 | 0.08 |
ENSDART00000062257
|
slc39a1
|
solute carrier family 39 (zinc transporter), member 1 |
| chr16_+_16824678 | 0.08 |
ENSDART00000172862
|
kcnj14
|
potassium inwardly-rectifying channel, subfamily J, member 14 |
| chr11_+_21050326 | 0.08 |
ENSDART00000065984
|
zgc:113307
|
zgc:113307 |
| chr22_+_19405517 | 0.08 |
ENSDART00000138245
ENSDART00000155144 |
si:dkey-78l4.2
|
si:dkey-78l4.2 |
| chr5_+_13385837 | 0.08 |
ENSDART00000191190
|
ccl19a.1
|
chemokine (C-C motif) ligand 19a, tandem duplicate 1 |
| chr15_-_29586747 | 0.08 |
ENSDART00000076749
|
samsn1a
|
SAM domain, SH3 domain and nuclear localisation signals 1a |
| chr25_+_13498188 | 0.07 |
ENSDART00000015710
|
snrkb
|
SNF related kinase b |
| chr12_+_19188542 | 0.07 |
ENSDART00000134726
ENSDART00000148011 ENSDART00000109541 |
cby1
|
chibby homolog 1 (Drosophila) |
| chr9_-_19699728 | 0.07 |
ENSDART00000166780
|
si:ch211-141e20.2
|
si:ch211-141e20.2 |
| chr19_+_40856807 | 0.07 |
ENSDART00000139083
|
gngt1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr15_-_5563551 | 0.07 |
ENSDART00000099520
|
atg16l2
|
ATG16 autophagy related 16-like 2 (S. cerevisiae) |
| chr9_+_14010823 | 0.07 |
ENSDART00000143837
|
si:ch211-67e16.3
|
si:ch211-67e16.3 |
| chr17_-_29192987 | 0.07 |
ENSDART00000164302
|
sptbn5
|
spectrin, beta, non-erythrocytic 5 |
| chr12_+_46740584 | 0.07 |
ENSDART00000171563
|
plaub
|
plasminogen activator, urokinase b |
| chr22_-_20289948 | 0.07 |
ENSDART00000132951
|
si:dkey-110c1.10
|
si:dkey-110c1.10 |
| chr2_+_30379650 | 0.06 |
ENSDART00000129542
|
crispld1b
|
cysteine-rich secretory protein LCCL domain containing 1b |
| chr25_-_15512819 | 0.06 |
ENSDART00000142684
|
si:dkeyp-67e1.2
|
si:dkeyp-67e1.2 |
| chr18_-_15771551 | 0.06 |
ENSDART00000130931
ENSDART00000154079 |
si:ch211-219a15.3
|
si:ch211-219a15.3 |
| chr4_-_34480599 | 0.06 |
ENSDART00000159602
ENSDART00000182248 |
si:dkeyp-51g9.4
si:ch211-64i20.3
|
si:dkeyp-51g9.4 si:ch211-64i20.3 |
| chr4_-_75899294 | 0.05 |
ENSDART00000157887
|
si:dkey-261j11.3
|
si:dkey-261j11.3 |
| chr4_-_60780423 | 0.05 |
ENSDART00000162632
|
si:dkey-254e13.6
|
si:dkey-254e13.6 |
| chr25_-_15504559 | 0.05 |
ENSDART00000139294
|
BX323543.5
|
|
| chr10_-_26738209 | 0.05 |
ENSDART00000188590
|
fgf13b
|
fibroblast growth factor 13b |
| chr4_+_64549970 | 0.05 |
ENSDART00000167846
|
si:ch211-223a21.3
|
si:ch211-223a21.3 |
| chr3_+_30968176 | 0.05 |
ENSDART00000186266
|
prf1.9
|
perforin 1.9 |
| chr4_+_51564997 | 0.05 |
ENSDART00000186119
|
si:dkey-165e24.1
|
si:dkey-165e24.1 |
| chr4_-_43616331 | 0.05 |
ENSDART00000183211
ENSDART00000150240 |
si:dkey-16p6.4
|
si:dkey-16p6.4 |
| chr22_+_19218733 | 0.05 |
ENSDART00000183212
ENSDART00000133595 |
si:dkey-21e2.7
|
si:dkey-21e2.7 |
| chr25_-_19486399 | 0.05 |
ENSDART00000155076
ENSDART00000156016 |
zgc:193812
|
zgc:193812 |
| chr4_+_54645654 | 0.05 |
ENSDART00000192864
|
si:ch211-227e10.1
|
si:ch211-227e10.1 |
| chr13_-_12602920 | 0.05 |
ENSDART00000102311
|
lrit3b
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 3b |
| chr3_+_60044780 | 0.05 |
ENSDART00000080437
|
zgc:113030
|
zgc:113030 |
| chr5_-_24270989 | 0.05 |
ENSDART00000146251
|
si:ch211-137i24.12
|
si:ch211-137i24.12 |
| chr18_-_36909773 | 0.04 |
ENSDART00000141694
|
si:ch211-160d20.5
|
si:ch211-160d20.5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of jund_batf
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.0 | GO:0043576 | respiratory gaseous exchange(GO:0007585) regulation of respiratory gaseous exchange(GO:0043576) |
| 0.6 | 1.9 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) |
| 0.3 | 1.1 | GO:0060547 | negative regulation of necroptotic process(GO:0060546) negative regulation of necrotic cell death(GO:0060547) |
| 0.2 | 0.7 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.2 | 0.7 | GO:0006600 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 0.2 | 0.6 | GO:0033335 | anal fin development(GO:0033335) |
| 0.2 | 0.9 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.2 | 1.3 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 0.7 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 0.1 | 0.7 | GO:0010872 | regulation of cholesterol esterification(GO:0010872) positive regulation of cholesterol esterification(GO:0010873) |
| 0.1 | 1.0 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.1 | 1.2 | GO:0043490 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.5 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.5 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 0.3 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.1 | 0.6 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.1 | 0.3 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 0.5 | GO:0060420 | regulation of heart growth(GO:0060420) |
| 0.1 | 0.2 | GO:0003408 | optic cup formation involved in camera-type eye development(GO:0003408) spinal cord association neuron specification(GO:0021519) |
| 0.1 | 0.2 | GO:0060137 | parturition(GO:0007567) neurohypophysis development(GO:0021985) maternal process involved in parturition(GO:0060137) |
| 0.1 | 1.2 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.1 | 0.2 | GO:0002631 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 0.1 | 0.4 | GO:0003352 | regulation of cilium movement(GO:0003352) |
| 0.1 | 0.2 | GO:1904088 | regulation of convergent extension involved in axis elongation(GO:1901232) positive regulation of epiboly involved in gastrulation with mouth forming second(GO:1904088) |
| 0.0 | 0.2 | GO:0015860 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.0 | 0.3 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.4 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.0 | 0.3 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 0.5 | GO:0014034 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.0 | 0.7 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.3 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 1.5 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.4 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.8 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.5 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.5 | GO:0009749 | response to glucose(GO:0009749) |
| 0.0 | 0.3 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.9 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.2 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.9 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.1 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.0 | 0.3 | GO:0060038 | cardiac muscle cell proliferation(GO:0060038) |
| 0.0 | 1.2 | GO:0006956 | complement activation(GO:0006956) |
| 0.0 | 0.6 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 0.1 | GO:0039694 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.3 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.2 | GO:0045807 | positive regulation of endocytosis(GO:0045807) |
| 0.0 | 0.0 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) semicircular canal fusion(GO:0060879) |
| 0.0 | 0.0 | GO:0006824 | cobalt ion transport(GO:0006824) cobalamin transport(GO:0015889) |
| 0.0 | 0.5 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 1.2 | GO:0050768 | negative regulation of neurogenesis(GO:0050768) |
| 0.0 | 0.5 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.1 | 0.5 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 0.7 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.9 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.7 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.8 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 1.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.1 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.4 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.0 | 0.9 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.0 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.2 | 0.9 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 1.0 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.7 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.1 | 1.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.6 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.1 | 1.2 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 0.5 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 1.2 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.1 | 1.1 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.1 | 0.2 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.1 | 1.3 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 0.3 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.1 | 0.2 | GO:0004990 | oxytocin receptor activity(GO:0004990) |
| 0.1 | 0.6 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 0.3 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 0.2 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.0 | 0.2 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.0 | 0.2 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.0 | 1.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.6 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.3 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.6 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.5 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.2 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.3 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.5 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.2 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.1 | GO:0048763 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.3 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.5 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.2 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.9 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.7 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.0 | 0.2 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.2 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.3 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.2 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.2 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.3 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.2 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.4 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 1.1 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.5 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.5 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.9 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.3 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.5 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.2 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.1 | 1.6 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 0.5 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.2 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 2.4 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.5 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.6 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.0 | 0.5 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.5 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.1 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |