Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
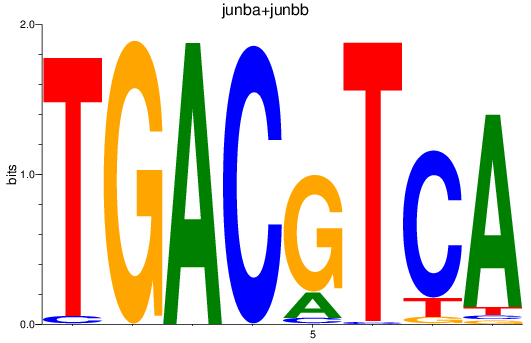
Results for junba+junbb
Z-value: 1.91
Transcription factors associated with junba+junbb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
junba
|
ENSDARG00000074378 | JunB proto-oncogene, AP-1 transcription factor subunit a |
|
junbb
|
ENSDARG00000104773 | JunB proto-oncogene, AP-1 transcription factor subunit b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| junba | dr11_v1_chr1_-_51734524_51734535 | 0.24 | 5.3e-01 | Click! |
| junbb | dr11_v1_chr3_-_7656059_7656059 | 0.15 | 7.0e-01 | Click! |
Activity profile of junba+junbb motif
Sorted Z-values of junba+junbb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_40992409 | 1.87 |
ENSDART00000151419
|
tgfa
|
transforming growth factor, alpha |
| chr25_-_23052707 | 1.54 |
ENSDART00000024633
|
dusp8a
|
dual specificity phosphatase 8a |
| chr21_-_4695583 | 1.49 |
ENSDART00000031425
|
zgc:55582
|
zgc:55582 |
| chr17_+_25187226 | 1.47 |
ENSDART00000148431
|
cln8
|
CLN8, transmembrane ER and ERGIC protein |
| chr23_+_19590598 | 1.46 |
ENSDART00000170149
|
slmapb
|
sarcolemma associated protein b |
| chr6_+_40992883 | 1.45 |
ENSDART00000076061
|
tgfa
|
transforming growth factor, alpha |
| chr17_-_12385308 | 1.40 |
ENSDART00000080927
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr23_+_19590006 | 1.39 |
ENSDART00000021231
|
slmapb
|
sarcolemma associated protein b |
| chr6_+_27667359 | 1.24 |
ENSDART00000159624
ENSDART00000049177 |
rab6ba
|
RAB6B, member RAS oncogene family a |
| chr10_-_34915886 | 1.24 |
ENSDART00000141201
ENSDART00000002166 |
ccna1
|
cyclin A1 |
| chr25_-_37084032 | 1.14 |
ENSDART00000025494
|
hprt1l
|
hypoxanthine phosphoribosyltransferase 1, like |
| chr22_+_24623936 | 1.10 |
ENSDART00000160924
|
mcoln2
|
mucolipin 2 |
| chr7_-_69429561 | 1.08 |
ENSDART00000127351
|
atxn1l
|
ataxin 1-like |
| chr16_-_383664 | 1.07 |
ENSDART00000051693
|
irx4a
|
iroquois homeobox 4a |
| chr2_-_37478418 | 1.06 |
ENSDART00000146103
|
dapk3
|
death-associated protein kinase 3 |
| chr4_-_16628801 | 1.03 |
ENSDART00000040708
ENSDART00000064009 |
caprin2
|
caprin family member 2 |
| chr15_-_34418525 | 1.02 |
ENSDART00000147582
|
agmo
|
alkylglycerol monooxygenase |
| chr12_-_10512911 | 1.02 |
ENSDART00000124562
ENSDART00000106163 |
zgc:152977
|
zgc:152977 |
| chr23_+_12160900 | 1.00 |
ENSDART00000136046
|
ppp1r3da
|
protein phosphatase 1, regulatory subunit 3Da |
| chr18_+_8917766 | 1.00 |
ENSDART00000145226
|
si:ch211-233h19.2
|
si:ch211-233h19.2 |
| chr18_+_910992 | 0.97 |
ENSDART00000161206
ENSDART00000167229 |
pkma
|
pyruvate kinase M1/2a |
| chr6_+_10333920 | 0.92 |
ENSDART00000151667
ENSDART00000151477 |
cobll1a
|
cordon-bleu WH2 repeat protein-like 1a |
| chr4_-_74998614 | 0.89 |
ENSDART00000162529
|
zgc:172139
|
zgc:172139 |
| chr20_-_36671660 | 0.87 |
ENSDART00000134819
|
slc5a6a
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6a |
| chr2_+_37227011 | 0.86 |
ENSDART00000126587
ENSDART00000084958 |
samd7
|
sterile alpha motif domain containing 7 |
| chr5_-_28016805 | 0.85 |
ENSDART00000078642
|
vps37b
|
vacuolar protein sorting 37 homolog B (S. cerevisiae) |
| chr8_+_23165749 | 0.84 |
ENSDART00000063057
|
dnajc5aa
|
DnaJ (Hsp40) homolog, subfamily C, member 5aa |
| chr19_-_874888 | 0.83 |
ENSDART00000007206
|
eomesa
|
eomesodermin homolog a |
| chr1_-_59240975 | 0.83 |
ENSDART00000166170
|
mvb12a
|
multivesicular body subunit 12A |
| chr14_-_41468892 | 0.82 |
ENSDART00000173099
ENSDART00000003170 |
mid1ip1l
|
MID1 interacting protein 1, like |
| chr18_-_21105968 | 0.80 |
ENSDART00000100791
|
si:dkey-12e7.1
|
si:dkey-12e7.1 |
| chr14_-_8903435 | 0.79 |
ENSDART00000160584
|
zgc:153681
|
zgc:153681 |
| chr7_-_20241346 | 0.78 |
ENSDART00000173619
ENSDART00000127699 |
si:ch73-335l21.4
|
si:ch73-335l21.4 |
| chr25_-_31763897 | 0.78 |
ENSDART00000041740
|
ubl7a
|
ubiquitin-like 7a (bone marrow stromal cell-derived) |
| chr21_-_43398122 | 0.77 |
ENSDART00000050533
|
ccni2
|
cyclin I family, member 2 |
| chr4_-_14915268 | 0.76 |
ENSDART00000067040
|
si:dkey-180p18.9
|
si:dkey-180p18.9 |
| chr4_-_73394385 | 0.75 |
ENSDART00000174205
ENSDART00000143417 |
zgc:162958
|
zgc:162958 |
| chr13_+_15816573 | 0.75 |
ENSDART00000137061
|
klc1a
|
kinesin light chain 1a |
| chr14_+_30285613 | 0.75 |
ENSDART00000173090
|
mtus1a
|
microtubule associated tumor suppressor 1a |
| chr16_-_42066523 | 0.74 |
ENSDART00000180538
ENSDART00000058620 |
zp3d.1
|
zona pellucida glycoprotein 3d tandem duplicate 1 |
| chr8_+_12951155 | 0.74 |
ENSDART00000081601
|
cept1a
|
choline/ethanolamine phosphotransferase 1a |
| chr12_-_22238004 | 0.73 |
ENSDART00000038310
|
ormdl3
|
ORMDL sphingolipid biosynthesis regulator 3 |
| chr23_+_44634187 | 0.72 |
ENSDART00000143688
|
si:ch73-265d7.2
|
si:ch73-265d7.2 |
| chr23_-_32157865 | 0.72 |
ENSDART00000000876
|
nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr7_-_48805181 | 0.72 |
ENSDART00000015884
|
mfge8a
|
milk fat globule-EGF factor 8 protein a |
| chr8_+_23174137 | 0.71 |
ENSDART00000189470
|
dnajc5aa
|
DnaJ (Hsp40) homolog, subfamily C, member 5aa |
| chr20_+_522457 | 0.70 |
ENSDART00000138585
|
nt5dc1
|
5'-nucleotidase domain containing 1 |
| chr9_-_46072805 | 0.70 |
ENSDART00000169682
|
hdac4
|
histone deacetylase 4 |
| chr15_+_25683069 | 0.70 |
ENSDART00000148190
|
hic1
|
hypermethylated in cancer 1 |
| chr12_-_25217217 | 0.70 |
ENSDART00000152931
|
kcng3
|
potassium voltage-gated channel, subfamily G, member 3 |
| chr2_+_25929619 | 0.70 |
ENSDART00000137746
|
slc7a14a
|
solute carrier family 7, member 14a |
| chr13_+_4505079 | 0.69 |
ENSDART00000144312
|
pde10a
|
phosphodiesterase 10A |
| chr6_+_12462079 | 0.69 |
ENSDART00000192029
ENSDART00000065385 |
nr4a2b
|
nuclear receptor subfamily 4, group A, member 2b |
| chr2_-_37477654 | 0.69 |
ENSDART00000193921
|
dapk3
|
death-associated protein kinase 3 |
| chr13_-_23956178 | 0.68 |
ENSDART00000133646
|
phactr2
|
phosphatase and actin regulator 2 |
| chr4_-_72643171 | 0.68 |
ENSDART00000130126
|
CABZ01054394.1
|
|
| chr3_-_49504023 | 0.68 |
ENSDART00000168108
|
prkacaa
|
protein kinase, cAMP-dependent, catalytic, alpha, genome duplicate a |
| chr10_+_5689510 | 0.68 |
ENSDART00000183217
ENSDART00000172632 |
pam
|
peptidylglycine alpha-amidating monooxygenase |
| chr9_-_18742704 | 0.67 |
ENSDART00000145401
|
tsc22d1
|
TSC22 domain family, member 1 |
| chr7_-_51749683 | 0.67 |
ENSDART00000083190
|
hdac8
|
histone deacetylase 8 |
| chr6_+_475264 | 0.67 |
ENSDART00000193615
|
LO017974.1
|
|
| chr6_-_52484566 | 0.67 |
ENSDART00000112146
|
fam83c
|
family with sequence similarity 83, member C |
| chr3_+_40255408 | 0.67 |
ENSDART00000074746
|
smcr8a
|
Smith-Magenis syndrome chromosome region, candidate 8a |
| chr21_-_43398457 | 0.66 |
ENSDART00000166530
|
ccni2
|
cyclin I family, member 2 |
| chr4_-_73562122 | 0.66 |
ENSDART00000174096
|
CU570689.2
|
|
| chr9_-_4598883 | 0.66 |
ENSDART00000171927
|
galnt13
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 13 |
| chr6_-_31348999 | 0.66 |
ENSDART00000153734
|
dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr5_+_36781732 | 0.66 |
ENSDART00000087191
|
mark4a
|
MAP/microtubule affinity-regulating kinase 4a |
| chr7_+_44593756 | 0.65 |
ENSDART00000125365
|
si:ch211-189a15.5
|
si:ch211-189a15.5 |
| chr18_+_16749091 | 0.64 |
ENSDART00000061265
|
rnf141
|
ring finger protein 141 |
| chr7_-_37555208 | 0.64 |
ENSDART00000148905
ENSDART00000150229 |
cylda
|
cylindromatosis (turban tumor syndrome), a |
| chr17_+_50657509 | 0.64 |
ENSDART00000179957
|
ddhd1a
|
DDHD domain containing 1a |
| chr25_+_3104959 | 0.64 |
ENSDART00000167130
|
rab3il1
|
RAB3A interacting protein (rabin3)-like 1 |
| chr13_-_23956361 | 0.63 |
ENSDART00000101150
|
phactr2
|
phosphatase and actin regulator 2 |
| chr25_+_33849647 | 0.63 |
ENSDART00000121449
|
roraa
|
RAR-related orphan receptor A, paralog a |
| chr15_+_20801253 | 0.63 |
ENSDART00000179387
|
aldh3a1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr16_+_27614989 | 0.63 |
ENSDART00000005625
|
glipr2l
|
GLI pathogenesis-related 2, like |
| chr2_-_55779927 | 0.63 |
ENSDART00000168579
|
CABZ01066725.1
|
|
| chr18_+_10784730 | 0.62 |
ENSDART00000028938
|
mical3a
|
microtubule associated monooxygenase, calponin and LIM domain containing 3a |
| chr8_+_25959940 | 0.62 |
ENSDART00000143011
ENSDART00000140626 |
si:dkey-72l14.4
|
si:dkey-72l14.4 |
| chr9_-_41507712 | 0.61 |
ENSDART00000135821
|
mfsd6b
|
major facilitator superfamily domain containing 6b |
| chr3_+_32403758 | 0.61 |
ENSDART00000156982
|
si:ch211-195b15.8
|
si:ch211-195b15.8 |
| chr6_+_28294113 | 0.61 |
ENSDART00000136898
|
lpp
|
LIM domain containing preferred translocation partner in lipoma |
| chr22_+_12366516 | 0.60 |
ENSDART00000157802
|
r3hdm1
|
R3H domain containing 1 |
| chr6_-_36795111 | 0.60 |
ENSDART00000160669
ENSDART00000104256 ENSDART00000187751 ENSDART00000161928 ENSDART00000183264 |
opa1
|
optic atrophy 1 (autosomal dominant) |
| chr15_-_28200049 | 0.60 |
ENSDART00000004200
|
sarm1
|
sterile alpha and TIR motif containing 1 |
| chr19_-_31707892 | 0.60 |
ENSDART00000088427
|
ripor2
|
RHO family interacting cell polarization regulator 2 |
| chr10_-_45379831 | 0.60 |
ENSDART00000186205
|
CABZ01117348.1
|
|
| chr7_+_13382852 | 0.59 |
ENSDART00000166318
|
dagla
|
diacylglycerol lipase, alpha |
| chr2_+_38002717 | 0.59 |
ENSDART00000139564
|
dcaf8
|
DDB1 and CUL4 associated factor 8 |
| chr22_+_30047245 | 0.59 |
ENSDART00000142857
ENSDART00000141247 ENSDART00000140015 ENSDART00000040538 |
add3a
|
adducin 3 (gamma) a |
| chr5_-_16425781 | 0.59 |
ENSDART00000185624
ENSDART00000180617 |
slc39a14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr3_-_13147310 | 0.58 |
ENSDART00000160840
|
prkar1b
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr1_-_46924801 | 0.58 |
ENSDART00000142560
|
pdxkb
|
pyridoxal (pyridoxine, vitamin B6) kinase b |
| chr6_-_60104628 | 0.58 |
ENSDART00000057463
ENSDART00000169188 |
pmepa1
|
prostate transmembrane protein, androgen induced 1 |
| chr6_-_40063359 | 0.58 |
ENSDART00000157119
ENSDART00000059636 ENSDART00000156385 |
chchd4b
|
coiled-coil-helix-coiled-coil-helix domain containing 4b |
| chr1_+_36772348 | 0.58 |
ENSDART00000109314
|
arhgap10
|
Rho GTPase activating protein 10 |
| chr19_-_30510930 | 0.58 |
ENSDART00000088760
ENSDART00000181043 |
bag6l
|
BCL2 associated athanogene 6, like |
| chr9_+_22003942 | 0.57 |
ENSDART00000091013
|
si:dkey-57a22.15
|
si:dkey-57a22.15 |
| chr9_-_18743012 | 0.57 |
ENSDART00000131626
|
tsc22d1
|
TSC22 domain family, member 1 |
| chr3_+_14463941 | 0.57 |
ENSDART00000170927
|
cnn1b
|
calponin 1, basic, smooth muscle, b |
| chr10_+_34426571 | 0.57 |
ENSDART00000144529
|
nbeaa
|
neurobeachin a |
| chr6_-_10988741 | 0.57 |
ENSDART00000090709
|
coq7
|
coenzyme Q7 homolog, ubiquinone (yeast) |
| chr4_+_9011825 | 0.57 |
ENSDART00000058007
|
samm50l
|
sorting and assembly machinery component 50 homolog, like |
| chr6_+_33537267 | 0.57 |
ENSDART00000040334
|
pik3r3b
|
phosphoinositide-3-kinase, regulatory subunit 3b (gamma) |
| chr8_+_29267093 | 0.56 |
ENSDART00000077647
|
grid2
|
glutamate receptor, ionotropic, delta 2 |
| chr25_-_27564205 | 0.56 |
ENSDART00000157319
|
hyal4
|
hyaluronoglucosaminidase 4 |
| chr2_-_16359042 | 0.55 |
ENSDART00000057216
|
arhgef4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr22_+_997838 | 0.55 |
ENSDART00000149743
|
pparda
|
peroxisome proliferator-activated receptor delta a |
| chr2_-_56635744 | 0.55 |
ENSDART00000167790
ENSDART00000168160 |
pip5k1cb
|
phosphatidylinositol-4-phosphate 5-kinase, type I, gamma b |
| chr19_+_22085925 | 0.55 |
ENSDART00000185636
|
atp9b
|
ATPase phospholipid transporting 9B |
| chr5_-_68782641 | 0.55 |
ENSDART00000141699
|
mepce
|
methylphosphate capping enzyme |
| chr21_+_25054420 | 0.55 |
ENSDART00000065132
|
zgc:171740
|
zgc:171740 |
| chr24_-_28229618 | 0.54 |
ENSDART00000145290
|
bcl2a
|
BCL2, apoptosis regulator a |
| chr8_+_44714336 | 0.54 |
ENSDART00000145801
|
elmod3
|
ELMO/CED-12 domain containing 3 |
| chr22_+_38914983 | 0.54 |
ENSDART00000085701
|
senp5
|
SUMO1/sentrin specific peptidase 5 |
| chr2_-_19354622 | 0.54 |
ENSDART00000168627
|
zfyve9a
|
zinc finger, FYVE domain containing 9a |
| chr20_-_44090624 | 0.54 |
ENSDART00000048978
ENSDART00000082283 ENSDART00000082276 |
runx2b
|
runt-related transcription factor 2b |
| chr9_-_17783574 | 0.54 |
ENSDART00000146706
|
vwa8
|
von Willebrand factor A domain containing 8 |
| chr2_+_32743807 | 0.54 |
ENSDART00000022909
|
klhl18
|
kelch-like family member 18 |
| chr5_-_9540641 | 0.54 |
ENSDART00000124384
ENSDART00000160079 |
gak
|
cyclin G associated kinase |
| chr15_-_8856391 | 0.53 |
ENSDART00000008273
|
rab4b
|
RAB4B, member RAS oncogene family |
| chr12_-_11560794 | 0.53 |
ENSDART00000149098
ENSDART00000169975 |
plekha1b
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1b |
| chr19_-_12648122 | 0.53 |
ENSDART00000151184
|
fam210aa
|
family with sequence similarity 210, member Aa |
| chr2_+_26060528 | 0.53 |
ENSDART00000058111
|
grin3ba
|
glutamate receptor, ionotropic, N-methyl-D-aspartate 3Ba |
| chr9_+_42095220 | 0.52 |
ENSDART00000148317
ENSDART00000134431 |
pcbp3
|
poly(rC) binding protein 3 |
| chr19_-_12648408 | 0.51 |
ENSDART00000103692
|
fam210aa
|
family with sequence similarity 210, member Aa |
| chr3_+_33367954 | 0.51 |
ENSDART00000103161
|
mpp2a
|
membrane protein, palmitoylated 2a (MAGUK p55 subfamily member 2) |
| chr22_+_786556 | 0.51 |
ENSDART00000125347
|
cry1bb
|
cryptochrome circadian clock 1bb |
| chr6_+_49901465 | 0.51 |
ENSDART00000023515
|
chmp4ba
|
charged multivesicular body protein 4Ba |
| chr6_+_36795225 | 0.51 |
ENSDART00000171504
|
si:ch73-29l19.1
|
si:ch73-29l19.1 |
| chr23_+_39281229 | 0.51 |
ENSDART00000180120
ENSDART00000193321 |
kcng1
|
potassium voltage-gated channel, subfamily G, member 1 |
| chr12_+_45200744 | 0.51 |
ENSDART00000098932
|
wbp2
|
WW domain binding protein 2 |
| chr3_-_15154871 | 0.50 |
ENSDART00000147704
|
si:dkey-282h22.5
|
si:dkey-282h22.5 |
| chr13_+_23988442 | 0.50 |
ENSDART00000010918
|
agt
|
angiotensinogen |
| chr2_-_42415902 | 0.50 |
ENSDART00000142489
|
slco5a1b
|
solute carrier organic anion transporter family member 5A1b |
| chr22_-_7168571 | 0.49 |
ENSDART00000165549
|
asic1b
|
acid-sensing (proton-gated) ion channel 1b |
| chr12_-_5455936 | 0.49 |
ENSDART00000109305
|
tbc1d12b
|
TBC1 domain family, member 12b |
| chr18_+_16330025 | 0.49 |
ENSDART00000142353
|
nts
|
neurotensin |
| chr17_+_47090497 | 0.49 |
ENSDART00000169038
ENSDART00000159292 |
zgc:103755
|
zgc:103755 |
| chr25_-_7686201 | 0.49 |
ENSDART00000157267
ENSDART00000155094 |
si:ch211-286c4.6
|
si:ch211-286c4.6 |
| chr7_-_50764714 | 0.49 |
ENSDART00000110283
|
iqgap1
|
IQ motif containing GTPase activating protein 1 |
| chr21_-_28640316 | 0.49 |
ENSDART00000128237
|
nrg2a
|
neuregulin 2a |
| chr2_-_37352514 | 0.49 |
ENSDART00000140498
ENSDART00000186422 |
skila
|
SKI-like proto-oncogene a |
| chr18_+_25752592 | 0.48 |
ENSDART00000111767
|
si:ch211-39k3.2
|
si:ch211-39k3.2 |
| chr10_+_7563755 | 0.48 |
ENSDART00000165877
|
purg
|
purine-rich element binding protein G |
| chr12_+_38807604 | 0.48 |
ENSDART00000155563
|
abca5
|
ATP-binding cassette, sub-family A (ABC1), member 5 |
| chr23_+_19655301 | 0.47 |
ENSDART00000104441
ENSDART00000135269 |
abhd6b
|
abhydrolase domain containing 6b |
| chr3_-_6709938 | 0.47 |
ENSDART00000172196
|
atg4db
|
autophagy related 4D, cysteine peptidase b |
| chr13_+_42309688 | 0.47 |
ENSDART00000158367
|
ide
|
insulin-degrading enzyme |
| chr11_-_40418975 | 0.47 |
ENSDART00000086296
|
trim62
|
tripartite motif containing 62 |
| chr21_+_5960443 | 0.47 |
ENSDART00000149689
|
mob1bb
|
MOB kinase activator 1Bb |
| chr20_-_53949798 | 0.47 |
ENSDART00000153435
|
ppp2r5cb
|
protein phosphatase 2, regulatory subunit B', gamma b |
| chr23_-_24542952 | 0.47 |
ENSDART00000088777
|
atp13a2
|
ATPase 13A2 |
| chr10_+_20392656 | 0.47 |
ENSDART00000160803
|
r3hcc1
|
R3H domain and coiled-coil containing 1 |
| chr11_-_17713987 | 0.46 |
ENSDART00000090401
|
fam19a4b
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A4b |
| chr9_+_45428041 | 0.46 |
ENSDART00000193087
|
adarb1b
|
adenosine deaminase, RNA-specific, B1b |
| chr11_-_7320211 | 0.46 |
ENSDART00000091664
|
apc2
|
adenomatosis polyposis coli 2 |
| chr17_+_50701748 | 0.46 |
ENSDART00000191938
ENSDART00000183220 ENSDART00000049464 |
fermt2
|
fermitin family member 2 |
| chr1_+_1941031 | 0.46 |
ENSDART00000110331
|
PTGFRN
|
si:ch211-132g1.7 |
| chr2_-_7800702 | 0.46 |
ENSDART00000146360
|
tbl1xr1b
|
transducin (beta)-like 1 X-linked receptor 1b |
| chr14_+_23929059 | 0.45 |
ENSDART00000159582
|
sh3rf2
|
SH3 domain containing ring finger 2 |
| chr4_-_21584158 | 0.45 |
ENSDART00000132923
|
ppp1r12a
|
protein phosphatase 1, regulatory subunit 12A |
| chr2_+_27830436 | 0.45 |
ENSDART00000182253
|
FO834800.2
|
|
| chr22_+_17261801 | 0.45 |
ENSDART00000192978
ENSDART00000193187 ENSDART00000179953 ENSDART00000134798 |
tdrd5
|
tudor domain containing 5 |
| chr19_+_41464870 | 0.45 |
ENSDART00000102778
|
dlx6a
|
distal-less homeobox 6a |
| chr6_+_112579 | 0.45 |
ENSDART00000034505
|
ap1m2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr16_+_41171949 | 0.45 |
ENSDART00000135294
|
nek11
|
NIMA-related kinase 11 |
| chr23_+_24973773 | 0.45 |
ENSDART00000047020
|
casp9
|
caspase 9, apoptosis-related cysteine peptidase |
| chr7_-_35515931 | 0.44 |
ENSDART00000193324
|
irx6a
|
iroquois homeobox 6a |
| chr16_+_14002944 | 0.44 |
ENSDART00000059926
|
si:dkey-85k15.7
|
si:dkey-85k15.7 |
| chr3_-_61494840 | 0.44 |
ENSDART00000101957
|
baiap2l1b
|
BAI1-associated protein 2-like 1b |
| chr24_-_7632187 | 0.44 |
ENSDART00000041714
|
atp6v0a1b
|
ATPase H+ transporting V0 subunit a1b |
| chr9_+_6998082 | 0.44 |
ENSDART00000092480
ENSDART00000135576 ENSDART00000188884 |
inpp4aa
|
inositol polyphosphate-4-phosphatase type I Aa |
| chr24_-_40901410 | 0.44 |
ENSDART00000170688
|
wdr48a
|
WD repeat domain 48a |
| chr13_+_24552254 | 0.44 |
ENSDART00000147907
|
lgalslb
|
lectin, galactoside-binding-like b |
| chr19_+_10396042 | 0.44 |
ENSDART00000028048
ENSDART00000151735 |
necap1
|
NECAP endocytosis associated 1 |
| chr17_-_8674208 | 0.43 |
ENSDART00000149201
|
ctbp2a
|
C-terminal binding protein 2a |
| chr2_+_9821757 | 0.43 |
ENSDART00000018408
ENSDART00000141227 ENSDART00000144681 ENSDART00000148227 |
anxa13l
|
annexin A13, like |
| chr20_+_3108597 | 0.43 |
ENSDART00000133435
|
CEP170B (1 of many)
|
si:ch73-212j7.1 |
| chr19_-_24136233 | 0.43 |
ENSDART00000143365
|
thap7
|
THAP domain containing 7 |
| chr8_+_48965767 | 0.43 |
ENSDART00000008058
|
aak1a
|
AP2 associated kinase 1a |
| chr13_+_534453 | 0.43 |
ENSDART00000147909
|
wu:fc17b08
|
wu:fc17b08 |
| chr7_-_15252540 | 0.43 |
ENSDART00000173072
ENSDART00000125258 |
si:dkey-172h23.2
|
si:dkey-172h23.2 |
| chr14_-_31080183 | 0.43 |
ENSDART00000173282
|
mbnl3
|
muscleblind-like splicing regulator 3 |
| chr7_+_34549377 | 0.43 |
ENSDART00000191814
|
fhod1
|
formin homology 2 domain containing 1 |
| chr21_-_25395223 | 0.43 |
ENSDART00000016219
|
ppme1
|
protein phosphatase methylesterase 1 |
| chr9_-_13871935 | 0.43 |
ENSDART00000146597
|
raph1a
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1a |
| chr7_+_34549198 | 0.42 |
ENSDART00000173784
|
fhod1
|
formin homology 2 domain containing 1 |
| chr10_-_35410518 | 0.42 |
ENSDART00000048430
|
gabrr3a
|
gamma-aminobutyric acid (GABA) A receptor, rho 3a |
| chr11_+_2710530 | 0.42 |
ENSDART00000132768
ENSDART00000030921 ENSDART00000040147 |
mapk14b
|
mitogen-activated protein kinase 14b |
| chr19_-_6840506 | 0.42 |
ENSDART00000081568
|
tcf19l
|
transcription factor 19 (SC1), like |
| chr1_-_11291324 | 0.42 |
ENSDART00000091205
|
sdk1b
|
sidekick cell adhesion molecule 1b |
| chr14_-_763744 | 0.42 |
ENSDART00000165856
|
trim35-27
|
tripartite motif containing 35-27 |
| chr6_+_11680726 | 0.42 |
ENSDART00000186732
|
calcrlb
|
calcitonin receptor-like b |
| chr4_+_9011448 | 0.42 |
ENSDART00000192357
|
samm50l
|
sorting and assembly machinery component 50 homolog, like |
| chr13_+_51710725 | 0.42 |
ENSDART00000163741
|
pwwp2b
|
PWWP domain containing 2B |
| chr22_+_28337429 | 0.42 |
ENSDART00000166177
|
impg2b
|
interphotoreceptor matrix proteoglycan 2b |
| chr19_-_24135824 | 0.41 |
ENSDART00000189505
ENSDART00000104087 |
thap7
|
THAP domain containing 7 |
| chr12_+_33038757 | 0.41 |
ENSDART00000153146
|
rbfox3a
|
RNA binding fox-1 homolog 3a |
Network of associatons between targets according to the STRING database.
First level regulatory network of junba+junbb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.6 | GO:2000273 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.4 | 1.1 | GO:0006178 | guanine salvage(GO:0006178) GMP salvage(GO:0032263) guanine biosynthetic process(GO:0046099) |
| 0.3 | 1.0 | GO:2000055 | positive regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000055) |
| 0.3 | 1.0 | GO:0045040 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.3 | 0.8 | GO:0060571 | invagination involved in gastrulation with mouth forming second(GO:0055109) morphogenesis of an epithelial fold(GO:0060571) |
| 0.3 | 0.8 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.2 | 1.2 | GO:0010754 | negative regulation of cGMP-mediated signaling(GO:0010754) |
| 0.2 | 0.7 | GO:0097623 | potassium ion export(GO:0071435) potassium ion export across plasma membrane(GO:0097623) |
| 0.2 | 0.9 | GO:0051182 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.2 | 1.0 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.2 | 0.6 | GO:0019677 | pyridine nucleotide catabolic process(GO:0019364) NAD catabolic process(GO:0019677) pyridine-containing compound catabolic process(GO:0072526) |
| 0.2 | 1.8 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.2 | 0.6 | GO:1990869 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) negative regulation of lymphocyte migration(GO:2000402) negative regulation of T cell migration(GO:2000405) |
| 0.2 | 1.2 | GO:0072318 | clathrin coat disassembly(GO:0072318) vesicle uncoating(GO:0072319) |
| 0.2 | 0.6 | GO:0033212 | iron assimilation(GO:0033212) |
| 0.2 | 0.6 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.2 | 0.5 | GO:1904871 | positive regulation of protein localization to nucleus(GO:1900182) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.2 | 0.7 | GO:1900060 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.2 | 0.7 | GO:0090299 | regulation of neural crest formation(GO:0090299) |
| 0.2 | 1.0 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.2 | 2.3 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.2 | 0.6 | GO:0002478 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) antigen processing and presentation of exogenous antigen(GO:0019884) antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.1 | 0.6 | GO:0071926 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 0.1 | 0.4 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.1 | 0.6 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.1 | 0.6 | GO:0010259 | multicellular organism aging(GO:0010259) |
| 0.1 | 1.0 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.1 | 0.7 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 0.6 | GO:1901842 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.8 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.1 | 0.3 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.1 | 0.7 | GO:0032528 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.1 | 0.5 | GO:0072574 | hepatocyte proliferation(GO:0072574) epithelial cell proliferation involved in liver morphogenesis(GO:0072575) |
| 0.1 | 0.3 | GO:0002631 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 0.1 | 0.3 | GO:0071586 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.1 | 0.6 | GO:0003151 | outflow tract morphogenesis(GO:0003151) |
| 0.1 | 0.7 | GO:0071326 | cellular response to carbohydrate stimulus(GO:0071322) cellular response to monosaccharide stimulus(GO:0071326) cellular response to hexose stimulus(GO:0071331) cellular response to glucose stimulus(GO:0071333) |
| 0.1 | 0.4 | GO:0052746 | inositol phosphorylation(GO:0052746) |
| 0.1 | 1.3 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.3 | GO:0003403 | optic vesicle formation(GO:0003403) |
| 0.1 | 0.6 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.1 | 0.5 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.1 | 0.4 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 0.6 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 0.5 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.1 | 0.7 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 1.2 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.1 | 0.3 | GO:0034729 | histone H3-K79 methylation(GO:0034729) regulation of transcription regulatory region DNA binding(GO:2000677) |
| 0.1 | 0.2 | GO:0055071 | manganese ion homeostasis(GO:0055071) |
| 0.1 | 0.3 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.1 | 0.5 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.1 | 0.2 | GO:1902410 | mitotic cytokinetic process(GO:1902410) mitotic cleavage furrow formation(GO:1903673) |
| 0.1 | 0.4 | GO:0099612 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.1 | 0.5 | GO:0098773 | ventricular trabecula myocardium morphogenesis(GO:0003222) trabecula formation(GO:0060343) heart trabecula formation(GO:0060347) skin epidermis development(GO:0098773) |
| 0.1 | 0.8 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.1 | 0.4 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.1 | 1.1 | GO:0048923 | posterior lateral line neuromast hair cell differentiation(GO:0048923) |
| 0.1 | 0.4 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.1 | 0.3 | GO:0080009 | 7-methylguanosine mRNA capping(GO:0006370) mRNA methylation(GO:0080009) |
| 0.1 | 0.7 | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0070059) |
| 0.1 | 0.7 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 0.6 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.1 | 0.3 | GO:0009794 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.1 | 0.4 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 1.1 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 | 0.6 | GO:0070309 | lens fiber cell development(GO:0070307) lens fiber cell morphogenesis(GO:0070309) |
| 0.1 | 0.2 | GO:0071788 | endoplasmic reticulum tubular network maintenance(GO:0071788) |
| 0.1 | 2.8 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.1 | 0.4 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.1 | 0.4 | GO:0001839 | neural plate morphogenesis(GO:0001839) |
| 0.1 | 0.2 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.0 | 0.1 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.0 | 0.2 | GO:0006266 | DNA ligation(GO:0006266) immunoglobulin V(D)J recombination(GO:0033152) DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 | 0.4 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.4 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.6 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.7 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 1.0 | GO:0005979 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.0 | 0.2 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.5 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.0 | 1.5 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 0.7 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.2 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 | 0.6 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.5 | GO:0039023 | pronephric duct morphogenesis(GO:0039023) |
| 0.0 | 0.5 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.6 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.5 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.0 | 0.8 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.4 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 1.7 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.1 | GO:0055107 | Golgi to secretory granule transport(GO:0055107) |
| 0.0 | 0.6 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.6 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.2 | GO:0035513 | oxidative RNA demethylation(GO:0035513) |
| 0.0 | 0.3 | GO:0044805 | late nucleophagy(GO:0044805) reticulophagy(GO:0061709) |
| 0.0 | 0.6 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.3 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.4 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.0 | 0.2 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.3 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.3 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.4 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.5 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 0.3 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.5 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 0.4 | GO:0045851 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.0 | 0.6 | GO:0035459 | cargo loading into vesicle(GO:0035459) |
| 0.0 | 0.3 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.7 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.4 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.0 | 0.3 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.4 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.3 | GO:0071715 | icosanoid transport(GO:0071715) fatty acid derivative transport(GO:1901571) |
| 0.0 | 0.2 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.1 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.0 | 0.3 | GO:0043490 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.5 | GO:0001990 | regulation of systemic arterial blood pressure by hormone(GO:0001990) |
| 0.0 | 0.1 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.7 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.3 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.0 | 0.3 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.4 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.5 | GO:0035141 | medial fin morphogenesis(GO:0035141) |
| 0.0 | 0.3 | GO:0090398 | cellular senescence(GO:0090398) |
| 0.0 | 0.5 | GO:1903845 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.2 | GO:0043097 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) pyrimidine nucleoside salvage(GO:0043097) UMP salvage(GO:0044206) |
| 0.0 | 0.3 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 1.5 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.0 | 0.3 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.3 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.3 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.1 | GO:0061438 | renal system vasculature morphogenesis(GO:0061438) kidney vasculature morphogenesis(GO:0061439) glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.0 | 0.8 | GO:0021587 | cerebellum morphogenesis(GO:0021587) |
| 0.0 | 0.2 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.9 | GO:0046890 | regulation of lipid biosynthetic process(GO:0046890) |
| 0.0 | 0.1 | GO:0050856 | regulation of T cell receptor signaling pathway(GO:0050856) |
| 0.0 | 0.1 | GO:0030262 | apoptotic DNA fragmentation(GO:0006309) cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.5 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.5 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.1 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.0 | 0.2 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 1.1 | GO:0032869 | cellular response to insulin stimulus(GO:0032869) |
| 0.0 | 0.5 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.8 | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage(GO:0008630) |
| 0.0 | 0.1 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.1 | GO:0090594 | inflammatory response to wounding(GO:0090594) |
| 0.0 | 0.4 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 0.3 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.0 | 0.2 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.4 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.3 | GO:0002042 | cell migration involved in sprouting angiogenesis(GO:0002042) |
| 0.0 | 0.4 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.4 | GO:0030537 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.0 | 0.1 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 0.5 | GO:0021549 | cerebellum development(GO:0021549) |
| 0.0 | 0.1 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.0 | 0.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.6 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:0003139 | secondary heart field specification(GO:0003139) |
| 0.0 | 0.2 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0098971 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.0 | 1.6 | GO:0043065 | positive regulation of cell death(GO:0010942) positive regulation of apoptotic process(GO:0043065) positive regulation of programmed cell death(GO:0043068) |
| 0.0 | 0.5 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.2 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.1 | GO:0070317 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.3 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 | 0.1 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.5 | GO:0045815 | positive regulation of gene expression, epigenetic(GO:0045815) |
| 0.0 | 0.2 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.2 | GO:0072554 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.0 | 0.6 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.0 | 0.3 | GO:1902749 | regulation of cell cycle G2/M phase transition(GO:1902749) |
| 0.0 | 0.3 | GO:0008214 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 | 0.3 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.2 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.3 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.0 | 0.3 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.0 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.1 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.9 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.2 | GO:0086010 | membrane depolarization(GO:0051899) membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.9 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.0 | 0.2 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 1.0 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.8 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.2 | GO:0046501 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.7 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.2 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.2 | 1.5 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.2 | 0.7 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.2 | 0.7 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.2 | 0.6 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.2 | 0.5 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 0.6 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.1 | 1.0 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.1 | 0.4 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.1 | 0.5 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 0.6 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 0.6 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.1 | 0.2 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.1 | 0.5 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 0.6 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.1 | 0.5 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 0.7 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 0.6 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.6 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.1 | 0.2 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.1 | 1.0 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.3 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.1 | 0.2 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.1 | 0.3 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.3 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 1.0 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.4 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.4 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.3 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.4 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.4 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.1 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.0 | 0.3 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.2 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 1.4 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.6 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 1.3 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.2 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.3 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 2.1 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.1 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.7 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 1.5 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.3 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.7 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.2 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.1 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.0 | 0.6 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.1 | GO:1990077 | primosome complex(GO:1990077) |
| 0.0 | 0.6 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.4 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.9 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.0 | 0.1 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) |
| 0.0 | 0.1 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.4 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.4 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.1 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 1.8 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 1.1 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.2 | GO:0008328 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.0 | 1.2 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 1.2 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 2.0 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.2 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.6 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.5 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.3 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 4.1 | GO:0098794 | postsynapse(GO:0098794) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0004422 | hypoxanthine phosphoribosyltransferase activity(GO:0004422) |
| 0.3 | 0.8 | GO:0001635 | adrenomedullin receptor activity(GO:0001605) calcitonin gene-related peptide receptor activity(GO:0001635) |
| 0.3 | 1.1 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.2 | 1.2 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.2 | 0.9 | GO:0008887 | glycerate kinase activity(GO:0008887) |
| 0.2 | 0.9 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.2 | 0.6 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.2 | 0.6 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) |
| 0.2 | 0.7 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.2 | 2.3 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.2 | 1.8 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.2 | 1.0 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.2 | 1.4 | GO:0097001 | sphingolipid binding(GO:0046625) ceramide binding(GO:0097001) |
| 0.2 | 0.8 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.2 | 3.6 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 0.7 | GO:0004307 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.1 | 1.1 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 1.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 1.7 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 0.8 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.1 | 0.6 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 0.3 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.1 | 0.2 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.1 | 1.0 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.1 | 0.7 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 0.5 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 0.7 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 0.3 | GO:0072571 | ADP-D-ribose binding(GO:0072570) mono-ADP-D-ribose binding(GO:0072571) |
| 0.1 | 1.0 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.6 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 0.3 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.1 | 0.3 | GO:0031151 | histone methyltransferase activity (H3-K79 specific)(GO:0031151) |
| 0.1 | 0.2 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.1 | 0.8 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 0.4 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 0.4 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.1 | 1.0 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 0.2 | GO:0055105 | ubiquitin-protein transferase inhibitor activity(GO:0055105) |
| 0.1 | 0.6 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 0.6 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 0.4 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.3 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.3 | GO:0005009 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.3 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.2 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.0 | 2.9 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 1.0 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.7 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.2 | GO:0035516 | oxidative DNA demethylase activity(GO:0035516) |
| 0.0 | 0.6 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.6 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.6 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 1.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.2 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.3 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.4 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.2 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.0 | 1.4 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.6 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.2 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.6 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 1.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.3 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.0 | 1.0 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 1.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.2 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 0.4 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.4 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.2 | GO:0015157 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.5 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.1 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.0 | 0.6 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.2 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 1.4 | GO:0004970 | ionotropic glutamate receptor activity(GO:0004970) |
| 0.0 | 0.2 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.1 | GO:0043878 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (non-phosphorylating) activity(GO:0043878) |
| 0.0 | 0.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.3 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.3 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.8 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 0.3 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.6 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.1 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.3 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.4 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.6 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.0 | 0.8 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.3 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.0 | 0.2 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.3 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.5 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.6 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.6 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.1 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.1 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
| 0.0 | 0.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.2 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.0 | 2.2 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.5 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.3 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.9 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.2 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.0 | GO:0015230 | folic acid transporter activity(GO:0008517) FAD transmembrane transporter activity(GO:0015230) |
| 0.0 | 0.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 0.2 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.2 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.8 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.1 | GO:0016917 | G-protein coupled GABA receptor activity(GO:0004965) GABA receptor activity(GO:0016917) |
| 0.0 | 0.3 | GO:0051119 | sugar transmembrane transporter activity(GO:0051119) |
| 0.0 | 0.1 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.3 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.7 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.3 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.6 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.6 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.2 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.2 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 1.1 | GO:0016298 | lipase activity(GO:0016298) |
| 0.0 | 0.5 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.6 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.9 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.7 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.2 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.2 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.0 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.0 | 0.3 | GO:0042287 | MHC protein binding(GO:0042287) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 0.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.8 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.7 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.7 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.5 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.9 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.8 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 1.3 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.5 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.6 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.4 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.2 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.2 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.1 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.2 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.2 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.2 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 1.3 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.2 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.4 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 1.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 1.2 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 2.0 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 0.9 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 0.6 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.3 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.6 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.7 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.2 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.5 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 1.7 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.3 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.3 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 1.4 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.2 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.6 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 1.1 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.4 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.1 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.0 | 0.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.3 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.1 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 0.4 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 0.2 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.9 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.3 | REACTOME PI METABOLISM | Genes involved in PI Metabolism |
| 0.0 | 0.5 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.2 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |