Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
Results for jun
Z-value: 0.84
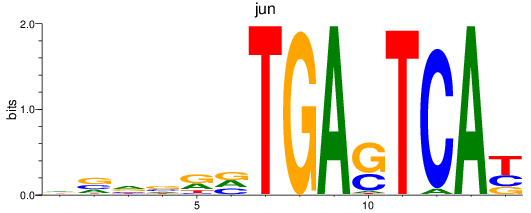
Transcription factors associated with jun
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
jun
|
ENSDARG00000043531 | Jun proto-oncogene, AP-1 transcription factor subunit |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| jun | dr11_v1_chr20_+_15552657_15552657 | 0.55 | 1.2e-01 | Click! |
Activity profile of jun motif
Sorted Z-values of jun motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_23403602 | 2.28 |
ENSDART00000159848
|
s100w
|
S100 calcium binding protein W |
| chr11_-_24681292 | 1.82 |
ENSDART00000089601
|
olfml3b
|
olfactomedin-like 3b |
| chr20_+_26880668 | 1.65 |
ENSDART00000077769
|
serpinb1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
| chr19_+_24488403 | 1.59 |
ENSDART00000052421
|
txnipa
|
thioredoxin interacting protein a |
| chr22_-_26595027 | 1.21 |
ENSDART00000184162
|
CABZ01072309.1
|
|
| chr23_-_21471022 | 1.18 |
ENSDART00000104206
|
her4.2
|
hairy-related 4, tandem duplicate 2 |
| chr1_-_59252973 | 1.13 |
ENSDART00000167061
|
si:ch1073-286c18.5
|
si:ch1073-286c18.5 |
| chr13_+_24280380 | 1.08 |
ENSDART00000184115
|
acta1b
|
actin, alpha 1b, skeletal muscle |
| chr23_+_35714574 | 1.07 |
ENSDART00000164616
|
tuba1c
|
tubulin, alpha 1c |
| chr5_+_37087583 | 1.05 |
ENSDART00000049900
|
tagln2
|
transgelin 2 |
| chr5_-_30615901 | 1.05 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr2_+_48288461 | 1.03 |
ENSDART00000141495
|
hes6
|
hes family bHLH transcription factor 6 |
| chr10_+_38610741 | 1.01 |
ENSDART00000126444
|
mmp13a
|
matrix metallopeptidase 13a |
| chr16_+_23398369 | 0.99 |
ENSDART00000037694
|
s100a10b
|
S100 calcium binding protein A10b |
| chr7_-_35432901 | 0.97 |
ENSDART00000026712
|
mmp2
|
matrix metallopeptidase 2 |
| chr21_+_25765734 | 0.96 |
ENSDART00000021664
|
cldnb
|
claudin b |
| chr1_-_25679339 | 0.95 |
ENSDART00000161703
ENSDART00000054230 |
fgg
|
fibrinogen gamma chain |
| chr23_+_36130883 | 0.95 |
ENSDART00000103132
|
hoxc4a
|
homeobox C4a |
| chr5_-_25582721 | 0.95 |
ENSDART00000123986
|
anxa1a
|
annexin A1a |
| chr20_-_26531850 | 0.95 |
ENSDART00000183317
ENSDART00000131994 |
mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1 like |
| chr16_-_21785261 | 0.91 |
ENSDART00000078858
|
si:ch73-86n18.1
|
si:ch73-86n18.1 |
| chr8_+_7359294 | 0.90 |
ENSDART00000121708
|
pcsk1nl
|
proprotein convertase subtilisin/kexin type 1 inhibitor, like |
| chr21_+_22845317 | 0.88 |
ENSDART00000065555
|
birc2
|
baculoviral IAP repeat containing 2 |
| chr20_-_26532167 | 0.87 |
ENSDART00000061914
|
mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1 like |
| chr11_+_13630107 | 0.86 |
ENSDART00000172220
|
si:ch211-1a19.3
|
si:ch211-1a19.3 |
| chr5_-_20195350 | 0.86 |
ENSDART00000139675
|
dao.1
|
D-amino-acid oxidase, tandem duplicate 1 |
| chr19_-_7420867 | 0.84 |
ENSDART00000081741
|
rab25a
|
RAB25, member RAS oncogene family a |
| chr19_+_7567763 | 0.81 |
ENSDART00000140411
|
s100a11
|
S100 calcium binding protein A11 |
| chr4_-_16412084 | 0.79 |
ENSDART00000188460
|
dcn
|
decorin |
| chr2_+_47581997 | 0.79 |
ENSDART00000112579
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr3_+_27027781 | 0.78 |
ENSDART00000065495
|
emp2
|
epithelial membrane protein 2 |
| chr16_+_42829735 | 0.77 |
ENSDART00000014956
|
polr3glb
|
polymerase (RNA) III (DNA directed) polypeptide G like b |
| chr16_-_45225520 | 0.77 |
ENSDART00000158855
|
fxyd1
|
FXYD domain containing ion transport regulator 1 (phospholemman) |
| chr6_-_49063085 | 0.76 |
ENSDART00000156124
|
si:ch211-105j21.9
|
si:ch211-105j21.9 |
| chr19_-_5332784 | 0.74 |
ENSDART00000010373
|
krt1-19d
|
keratin, type 1, gene 19d |
| chr2_-_2020044 | 0.74 |
ENSDART00000024135
|
tubb2
|
tubulin, beta 2A class IIa |
| chr1_-_25177086 | 0.73 |
ENSDART00000144711
ENSDART00000177225 |
tmem154
|
transmembrane protein 154 |
| chr7_-_28696556 | 0.73 |
ENSDART00000148822
|
adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr2_+_47582681 | 0.73 |
ENSDART00000187579
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr9_-_48370645 | 0.73 |
ENSDART00000140185
|
col28a2a
|
collagen, type XXVIII, alpha 2a |
| chr4_-_6809323 | 0.73 |
ENSDART00000099467
|
ifrd1
|
interferon-related developmental regulator 1 |
| chr1_+_12766351 | 0.72 |
ENSDART00000165785
|
pcdh10a
|
protocadherin 10a |
| chr11_+_6116503 | 0.70 |
ENSDART00000176170
|
nr2f6b
|
nuclear receptor subfamily 2, group F, member 6b |
| chr13_-_16222388 | 0.65 |
ENSDART00000182861
|
zgc:110045
|
zgc:110045 |
| chr2_+_30379650 | 0.64 |
ENSDART00000129542
|
crispld1b
|
cysteine-rich secretory protein LCCL domain containing 1b |
| chr1_-_5746030 | 0.64 |
ENSDART00000150863
|
nrp2a
|
neuropilin 2a |
| chr20_-_43771871 | 0.63 |
ENSDART00000153304
|
matn3a
|
matrilin 3a |
| chr2_+_35612621 | 0.63 |
ENSDART00000143082
|
si:dkey-4i23.5
|
si:dkey-4i23.5 |
| chr7_+_25033924 | 0.62 |
ENSDART00000170873
|
sb:cb1058
|
sb:cb1058 |
| chr8_+_39511932 | 0.62 |
ENSDART00000113511
|
lzts1
|
leucine zipper, putative tumor suppressor 1 |
| chr6_-_18976168 | 0.62 |
ENSDART00000170039
|
sept9b
|
septin 9b |
| chr10_+_37145007 | 0.62 |
ENSDART00000131777
|
cuedc1a
|
CUE domain containing 1a |
| chr11_-_18253111 | 0.61 |
ENSDART00000125984
|
mustn1b
|
musculoskeletal, embryonic nuclear protein 1b |
| chr8_+_41647539 | 0.61 |
ENSDART00000136492
ENSDART00000138799 ENSDART00000134404 |
si:ch211-158d24.4
|
si:ch211-158d24.4 |
| chr5_-_55623443 | 0.61 |
ENSDART00000005671
ENSDART00000176341 |
hnrpkl
|
heterogeneous nuclear ribonucleoprotein K, like |
| chr20_+_34915945 | 0.60 |
ENSDART00000153064
|
snap25a
|
synaptosomal-associated protein, 25a |
| chr13_+_829585 | 0.59 |
ENSDART00000029051
|
gsta.2
|
glutathione S-transferase, alpha tandem duplicate 2 |
| chr3_-_26017592 | 0.59 |
ENSDART00000030890
|
hmox1a
|
heme oxygenase 1a |
| chr11_+_13629528 | 0.59 |
ENSDART00000186509
|
si:ch211-1a19.3
|
si:ch211-1a19.3 |
| chr8_+_39634114 | 0.59 |
ENSDART00000144293
|
msi1
|
musashi RNA-binding protein 1 |
| chr21_-_30545121 | 0.59 |
ENSDART00000019199
|
rab39ba
|
RAB39B, member RAS oncogene family a |
| chr10_+_22771176 | 0.59 |
ENSDART00000192046
|
tmem88a
|
transmembrane protein 88 a |
| chr3_-_26017831 | 0.58 |
ENSDART00000179982
|
hmox1a
|
heme oxygenase 1a |
| chr12_+_25945560 | 0.58 |
ENSDART00000109799
|
mmrn2b
|
multimerin 2b |
| chr23_+_9067131 | 0.56 |
ENSDART00000144533
|
ccm2l
|
cerebral cavernous malformation 2-like |
| chr13_+_50368668 | 0.56 |
ENSDART00000127610
|
dnajc12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr6_+_56141852 | 0.56 |
ENSDART00000149665
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr11_+_6116096 | 0.54 |
ENSDART00000159680
|
nr2f6b
|
nuclear receptor subfamily 2, group F, member 6b |
| chr13_+_23988442 | 0.54 |
ENSDART00000010918
|
agt
|
angiotensinogen |
| chr16_+_42830152 | 0.54 |
ENSDART00000159730
|
polr3glb
|
polymerase (RNA) III (DNA directed) polypeptide G like b |
| chr4_+_16715267 | 0.52 |
ENSDART00000143849
|
pkp2
|
plakophilin 2 |
| chr9_-_710896 | 0.52 |
ENSDART00000180478
|
ndufb3
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3 |
| chr5_+_15819651 | 0.51 |
ENSDART00000081230
ENSDART00000186969 ENSDART00000134206 |
hspb8
|
heat shock protein b8 |
| chr19_-_103289 | 0.51 |
ENSDART00000143118
|
adgrb1b
|
adhesion G protein-coupled receptor B1b |
| chr14_+_33882973 | 0.51 |
ENSDART00000019396
|
clic2
|
chloride intracellular channel 2 |
| chr7_-_8470860 | 0.48 |
ENSDART00000172793
|
loc564481
|
hypothetical protein LOC564481 |
| chr7_-_52963493 | 0.48 |
ENSDART00000052029
|
cart3
|
cocaine- and amphetamine-regulated transcript 3 |
| chr13_+_35339182 | 0.48 |
ENSDART00000019323
|
jag1b
|
jagged 1b |
| chr2_+_47582488 | 0.48 |
ENSDART00000149967
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr5_+_37785152 | 0.48 |
ENSDART00000053511
ENSDART00000189812 |
myo1ca
|
myosin Ic, paralog a |
| chr3_+_20156956 | 0.48 |
ENSDART00000125281
|
ngfra
|
nerve growth factor receptor a (TNFR superfamily, member 16) |
| chr9_-_2594410 | 0.48 |
ENSDART00000188306
ENSDART00000164276 |
sp9
|
sp9 transcription factor |
| chr10_+_38593645 | 0.48 |
ENSDART00000011573
|
mmp13a
|
matrix metallopeptidase 13a |
| chr8_+_32406885 | 0.47 |
ENSDART00000167600
|
epgn
|
epithelial mitogen homolog (mouse) |
| chr20_-_42972599 | 0.47 |
ENSDART00000100751
|
pomcb
|
proopiomelanocortin b |
| chr3_-_39648021 | 0.47 |
ENSDART00000055171
|
grapa
|
GRB2-related adaptor protein a |
| chr5_-_26879302 | 0.46 |
ENSDART00000098571
ENSDART00000139086 |
zgc:64051
|
zgc:64051 |
| chr14_+_33722950 | 0.46 |
ENSDART00000075312
|
apln
|
apelin |
| chr14_-_17588345 | 0.46 |
ENSDART00000143486
|
selenot2
|
selenoprotein T, 2 |
| chr24_+_20575259 | 0.46 |
ENSDART00000010488
|
klhl40b
|
kelch-like family member 40b |
| chr3_+_21189766 | 0.44 |
ENSDART00000078807
|
zgc:123295
|
zgc:123295 |
| chr19_-_41404870 | 0.44 |
ENSDART00000167772
|
sem1
|
SEM1, 26S proteasome complex subunit |
| chr16_-_48673938 | 0.44 |
ENSDART00000156969
|
notchl
|
notch homolog, like |
| chr17_-_7861219 | 0.43 |
ENSDART00000148604
|
syne1b
|
spectrin repeat containing, nuclear envelope 1b |
| chr16_+_50006872 | 0.43 |
ENSDART00000157100
|
ube2e2
|
ubiquitin-conjugating enzyme E2E 2 |
| chr5_-_34616599 | 0.43 |
ENSDART00000050271
ENSDART00000097975 |
hexb
|
hexosaminidase B (beta polypeptide) |
| chr7_+_69019851 | 0.42 |
ENSDART00000162891
|
CABZ01057488.1
|
|
| chr1_-_22861348 | 0.42 |
ENSDART00000139412
|
SMIM18
|
si:dkey-92j12.6 |
| chr1_+_59090743 | 0.42 |
ENSDART00000100199
|
mfap4
|
microfibril associated protein 4 |
| chr1_+_41849152 | 0.41 |
ENSDART00000053685
|
smox
|
spermine oxidase |
| chr9_-_6372535 | 0.41 |
ENSDART00000149189
|
ecrg4a
|
esophageal cancer related gene 4a |
| chr19_-_38539670 | 0.41 |
ENSDART00000136775
|
col16a1
|
collagen, type XVI, alpha 1 |
| chr19_+_22216778 | 0.40 |
ENSDART00000052521
|
nfatc1
|
nuclear factor of activated T cells 1 |
| chr23_-_1017605 | 0.40 |
ENSDART00000138290
|
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr19_-_42573219 | 0.40 |
ENSDART00000126021
ENSDART00000133695 ENSDART00000131558 |
zgc:103438
|
zgc:103438 |
| chr8_-_25338709 | 0.40 |
ENSDART00000131616
|
atp5pb
|
ATP synthase peripheral stalk-membrane subunit b |
| chr1_+_59090972 | 0.39 |
ENSDART00000171497
|
mfap4
|
microfibril associated protein 4 |
| chr4_-_73787702 | 0.39 |
ENSDART00000136328
ENSDART00000150546 |
si:dkey-262g12.3
|
si:dkey-262g12.3 |
| chr25_-_22191733 | 0.39 |
ENSDART00000067478
|
pkp3a
|
plakophilin 3a |
| chr25_-_37331513 | 0.39 |
ENSDART00000111862
|
ldlrad3
|
low density lipoprotein receptor class A domain containing 3 |
| chr20_-_40319890 | 0.39 |
ENSDART00000075112
|
clvs2
|
clavesin 2 |
| chr17_-_52091999 | 0.38 |
ENSDART00000019766
|
tgfb3
|
transforming growth factor, beta 3 |
| chr15_-_46779934 | 0.38 |
ENSDART00000085136
|
clcn2c
|
chloride channel 2c |
| chr7_+_23875269 | 0.38 |
ENSDART00000101406
|
rab39bb
|
RAB39B, member RAS oncogene family b |
| chr20_-_25626198 | 0.37 |
ENSDART00000126716
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr10_+_4499943 | 0.37 |
ENSDART00000125299
|
plk2a
|
polo-like kinase 2a (Drosophila) |
| chr25_+_21324588 | 0.37 |
ENSDART00000151842
|
lrrn3a
|
leucine rich repeat neuronal 3a |
| chr7_+_48460239 | 0.37 |
ENSDART00000052113
|
lingo1b
|
leucine rich repeat and Ig domain containing 1b |
| chr23_+_42336084 | 0.37 |
ENSDART00000158959
ENSDART00000161812 |
cyp2aa7
cyp2aa8
|
cytochrome P450, family 2, subfamily AA, polypeptide 7 cytochrome P450, family 2, subfamily AA, polypeptide 8 |
| chr10_-_3295197 | 0.37 |
ENSDART00000109131
|
slc25a1b
|
slc25a1 solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1b |
| chr3_+_37574885 | 0.36 |
ENSDART00000055225
|
wnt9b
|
wingless-type MMTV integration site family, member 9B |
| chr7_-_17690983 | 0.36 |
ENSDART00000009367
|
map4k2
|
mitogen-activated protein kinase kinase kinase kinase 2 |
| chr16_-_36834505 | 0.36 |
ENSDART00000141275
ENSDART00000139588 ENSDART00000041993 |
pnp4b
|
purine nucleoside phosphorylase 4b |
| chr3_-_48259289 | 0.36 |
ENSDART00000160717
|
znf750
|
zinc finger protein 750 |
| chr7_+_20298333 | 0.35 |
ENSDART00000023089
ENSDART00000131019 |
acadvl
|
acyl-CoA dehydrogenase very long chain |
| chr23_-_7797207 | 0.35 |
ENSDART00000181611
|
myt1b
|
myelin transcription factor 1b |
| chr7_+_34492744 | 0.35 |
ENSDART00000109635
ENSDART00000173844 |
calml4a
|
calmodulin-like 4a |
| chr7_-_22941472 | 0.35 |
ENSDART00000190334
|
tnfsf10l
|
TNF superfamily member 10, like |
| chr14_+_49382180 | 0.34 |
ENSDART00000158329
|
tnip1
|
TNFAIP3 interacting protein 1 |
| chr22_+_16497670 | 0.34 |
ENSDART00000014330
|
ier5
|
immediate early response 5 |
| chr14_+_30279391 | 0.34 |
ENSDART00000172794
|
fgl1
|
fibrinogen-like 1 |
| chr23_-_4915118 | 0.34 |
ENSDART00000060714
|
atp6ap1a
|
ATPase H+ transporting accessory protein 1a |
| chr10_-_27566481 | 0.34 |
ENSDART00000078920
|
auts2a
|
autism susceptibility candidate 2a |
| chr23_+_24124684 | 0.34 |
ENSDART00000144478
|
si:dkey-21o19.2
|
si:dkey-21o19.2 |
| chr9_+_41024973 | 0.34 |
ENSDART00000014660
ENSDART00000144467 |
ormdl1
|
ORMDL sphingolipid biosynthesis regulator 1 |
| chr18_+_16125852 | 0.34 |
ENSDART00000061106
|
bhlhe41
|
basic helix-loop-helix family, member e41 |
| chr22_-_11724563 | 0.33 |
ENSDART00000190796
ENSDART00000184744 |
krt222
|
keratin 222 |
| chr11_-_10770053 | 0.33 |
ENSDART00000179213
|
slc4a10a
|
solute carrier family 4, sodium bicarbonate transporter, member 10a |
| chr15_+_9327252 | 0.33 |
ENSDART00000144381
|
sgcg
|
sarcoglycan, gamma |
| chr6_-_31224563 | 0.32 |
ENSDART00000104616
|
lepr
|
leptin receptor |
| chr19_+_37120491 | 0.32 |
ENSDART00000032341
|
pef1
|
penta-EF-hand domain containing 1 |
| chr7_+_69470442 | 0.32 |
ENSDART00000189593
|
gabarapb
|
GABA(A) receptor-associated protein b |
| chr16_-_27628994 | 0.32 |
ENSDART00000157407
|
nacad
|
NAC alpha domain containing |
| chr21_-_42100471 | 0.32 |
ENSDART00000166148
|
gabra1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr10_-_43928937 | 0.32 |
ENSDART00000168014
ENSDART00000086220 |
mctp1b
|
multiple C2 domains, transmembrane 1b |
| chr13_-_21701323 | 0.32 |
ENSDART00000164112
|
si:dkey-191g9.7
|
si:dkey-191g9.7 |
| chr7_+_20587506 | 0.31 |
ENSDART00000172416
ENSDART00000170633 |
si:dkey-19b23.7
|
si:dkey-19b23.7 |
| chr5_+_42280372 | 0.31 |
ENSDART00000142855
|
tbx6l
|
T-box 6, like |
| chr13_-_17729474 | 0.31 |
ENSDART00000013011
|
vdac2
|
voltage-dependent anion channel 2 |
| chr1_+_59073436 | 0.31 |
ENSDART00000161642
|
MFAP4 (1 of many)
|
si:zfos-2330d3.3 |
| chr17_+_45413324 | 0.30 |
ENSDART00000124911
|
ezra
|
ezrin a |
| chr4_+_1600034 | 0.30 |
ENSDART00000146779
|
slc38a2
|
solute carrier family 38, member 2 |
| chr15_-_42736433 | 0.30 |
ENSDART00000154379
|
si:ch211-181d7.1
|
si:ch211-181d7.1 |
| chr3_+_22375596 | 0.30 |
ENSDART00000188243
ENSDART00000181506 |
arhgap27l
|
Rho GTPase activating protein 27, like |
| chr12_+_20587179 | 0.29 |
ENSDART00000170127
|
arsg
|
arylsulfatase G |
| chr13_-_37122217 | 0.29 |
ENSDART00000133242
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr1_+_7988052 | 0.29 |
ENSDART00000167552
|
CR855320.2
|
|
| chr18_-_26785861 | 0.29 |
ENSDART00000098361
|
nmba
|
neuromedin Ba |
| chr16_+_10972733 | 0.29 |
ENSDART00000049323
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr22_-_32392252 | 0.28 |
ENSDART00000148681
ENSDART00000137945 |
pcbp4
|
poly(rC) binding protein 4 |
| chr13_-_15982707 | 0.28 |
ENSDART00000186911
ENSDART00000181072 |
ikzf1
|
IKAROS family zinc finger 1 (Ikaros) |
| chr23_-_28239750 | 0.28 |
ENSDART00000003548
|
znf385a
|
zinc finger protein 385A |
| chr1_-_35694978 | 0.28 |
ENSDART00000136157
|
si:dkey-27h10.2
|
si:dkey-27h10.2 |
| chr11_+_36158134 | 0.28 |
ENSDART00000189827
ENSDART00000163330 |
grm2b
|
glutamate receptor, metabotropic 2b |
| chr2_-_985417 | 0.27 |
ENSDART00000140540
|
si:ch211-241e1.3
|
si:ch211-241e1.3 |
| chr23_-_1017428 | 0.27 |
ENSDART00000110588
ENSDART00000183158 |
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr1_-_55116453 | 0.27 |
ENSDART00000142348
|
sertad2a
|
SERTA domain containing 2a |
| chr6_-_24143923 | 0.27 |
ENSDART00000157948
|
si:ch73-389b16.1
|
si:ch73-389b16.1 |
| chr4_-_13902188 | 0.27 |
ENSDART00000032805
|
gxylt1b
|
glucoside xylosyltransferase 1b |
| chr22_-_10110959 | 0.27 |
ENSDART00000031005
ENSDART00000147580 |
gls2b
|
glutaminase 2b (liver, mitochondrial) |
| chr10_+_31222433 | 0.26 |
ENSDART00000185080
|
tmem218
|
transmembrane protein 218 |
| chr1_+_14253118 | 0.26 |
ENSDART00000161996
|
cxcl8a
|
chemokine (C-X-C motif) ligand 8a |
| chr10_+_39263827 | 0.26 |
ENSDART00000172509
|
foxred1
|
FAD-dependent oxidoreductase domain containing 1 |
| chr5_-_46896541 | 0.26 |
ENSDART00000133240
|
edil3a
|
EGF-like repeats and discoidin I-like domains 3a |
| chr23_-_29376859 | 0.26 |
ENSDART00000146411
|
sst6
|
somatostatin 6 |
| chr6_-_43449013 | 0.25 |
ENSDART00000122423
|
eevs
|
2-epi-5-epi-valiolone synthase |
| chr25_-_8030425 | 0.25 |
ENSDART00000014964
|
camk1db
|
calcium/calmodulin-dependent protein kinase 1Db |
| chr3_+_7617353 | 0.25 |
ENSDART00000165551
|
zgc:109949
|
zgc:109949 |
| chr18_+_3052408 | 0.25 |
ENSDART00000181563
|
kctd14
|
potassium channel tetramerization domain containing 14 |
| chr5_-_48260145 | 0.25 |
ENSDART00000044083
ENSDART00000163250 ENSDART00000135911 |
mef2cb
|
myocyte enhancer factor 2cb |
| chr7_+_50395856 | 0.25 |
ENSDART00000032324
|
hddc3
|
HD domain containing 3 |
| chr2_-_32558795 | 0.25 |
ENSDART00000140026
|
smarcd3a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3a |
| chr10_+_31222656 | 0.24 |
ENSDART00000140988
ENSDART00000143387 |
tmem218
|
transmembrane protein 218 |
| chr8_-_23573084 | 0.24 |
ENSDART00000139084
|
wasb
|
Wiskott-Aldrich syndrome (eczema-thrombocytopenia) b |
| chr9_-_42418470 | 0.24 |
ENSDART00000144353
|
calcrla
|
calcitonin receptor-like a |
| chr6_+_52350443 | 0.23 |
ENSDART00000151612
ENSDART00000151349 |
si:ch211-239j9.1
|
si:ch211-239j9.1 |
| chr20_+_26095530 | 0.23 |
ENSDART00000139350
|
syne1a
|
spectrin repeat containing, nuclear envelope 1a |
| chr3_-_58189429 | 0.23 |
ENSDART00000156092
|
si:ch211-256e16.11
|
si:ch211-256e16.11 |
| chr3_-_32320537 | 0.23 |
ENSDART00000113550
ENSDART00000168483 |
si:dkey-16p21.7
|
si:dkey-16p21.7 |
| chr1_+_56447107 | 0.23 |
ENSDART00000091924
|
CABZ01059392.1
|
|
| chr5_+_9037650 | 0.23 |
ENSDART00000158226
|
si:ch211-155m12.1
|
si:ch211-155m12.1 |
| chr15_-_31177324 | 0.23 |
ENSDART00000008854
|
wsb1
|
WD repeat and SOCS box containing 1 |
| chr23_+_16814968 | 0.23 |
ENSDART00000161299
|
zgc:100832
|
zgc:100832 |
| chr1_+_45323142 | 0.23 |
ENSDART00000132210
|
emp1
|
epithelial membrane protein 1 |
| chr20_-_35578435 | 0.23 |
ENSDART00000142444
|
adgrf6
|
adhesion G protein-coupled receptor F6 |
| chr15_+_29123031 | 0.23 |
ENSDART00000133988
ENSDART00000060030 |
zgc:101731
|
zgc:101731 |
| chr8_-_38105053 | 0.22 |
ENSDART00000131546
|
adgra2
|
adhesion G protein-coupled receptor A2 |
| chr19_+_48018464 | 0.22 |
ENSDART00000172307
ENSDART00000163848 |
UBE2M
|
si:ch1073-205c8.3 |
| chr23_-_437467 | 0.22 |
ENSDART00000192106
|
tspan2b
|
tetraspanin 2b |
| chr1_-_58913813 | 0.22 |
ENSDART00000056494
|
zgc:171687
|
zgc:171687 |
Network of associatons between targets according to the STRING database.
First level regulatory network of jun
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0043576 | respiratory gaseous exchange(GO:0007585) regulation of respiratory gaseous exchange(GO:0043576) |
| 0.3 | 0.8 | GO:0001977 | renal system process involved in regulation of blood volume(GO:0001977) renal system process involved in regulation of systemic arterial blood pressure(GO:0003071) regulation of glomerular filtration(GO:0003093) |
| 0.2 | 0.9 | GO:0060547 | negative regulation of necroptotic process(GO:0060546) negative regulation of necrotic cell death(GO:0060547) |
| 0.2 | 0.9 | GO:0009080 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.2 | 0.6 | GO:1905208 | negative regulation of striated muscle cell differentiation(GO:0051154) negative regulation of cardiac muscle tissue development(GO:0055026) cardiac muscle cell fate commitment(GO:0060923) canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) negative regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901296) negative regulation of cardiocyte differentiation(GO:1905208) negative regulation of cardiac muscle cell differentiation(GO:2000726) |
| 0.2 | 0.9 | GO:0032663 | neutrophil apoptotic process(GO:0001781) regulation of T-helper 1 type immune response(GO:0002825) positive regulation of T-helper 1 type immune response(GO:0002827) negative regulation of type 2 immune response(GO:0002829) inflammatory cell apoptotic process(GO:0006925) regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of phospholipase A2 activity(GO:0032429) interleukin-2 production(GO:0032623) regulation of interleukin-2 production(GO:0032663) positive regulation of interleukin-2 production(GO:0032743) myeloid cell apoptotic process(GO:0033028) regulation of neutrophil apoptotic process(GO:0033029) positive regulation of neutrophil apoptotic process(GO:0033031) regulation of myeloid cell apoptotic process(GO:0033032) positive regulation of myeloid cell apoptotic process(GO:0033034) T-helper 1 type immune response(GO:0042088) positive regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043372) T-helper 1 cell differentiation(GO:0045063) positive regulation of T-helper cell differentiation(GO:0045624) regulation of T-helper 1 cell differentiation(GO:0045625) positive regulation of T-helper 1 cell differentiation(GO:0045627) regulation of T-helper 2 cell differentiation(GO:0045628) negative regulation of T-helper 2 cell differentiation(GO:0045629) positive regulation of fatty acid biosynthetic process(GO:0045723) positive regulation of alpha-beta T cell differentiation(GO:0046638) neutrophil clearance(GO:0097350) negative regulation of phospholipase A2 activity(GO:1900138) positive regulation of CD4-positive, alpha-beta T cell activation(GO:2000516) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.2 | 0.5 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.1 | 0.7 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.1 | 0.4 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.1 | 0.5 | GO:0090387 | phagosome maturation involved in apoptotic cell clearance(GO:0090386) phagolysosome assembly involved in apoptotic cell clearance(GO:0090387) |
| 0.1 | 0.7 | GO:0070445 | regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.1 | 1.8 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.1 | 0.4 | GO:1990791 | dorsal root ganglion development(GO:1990791) |
| 0.1 | 0.5 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.1 | 0.4 | GO:0006843 | mitochondrial citrate transport(GO:0006843) |
| 0.1 | 0.3 | GO:0002432 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 0.1 | 0.3 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.1 | 0.3 | GO:2000303 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.1 | 2.5 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.1 | 0.5 | GO:0090133 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.1 | 1.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.5 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.1 | 1.0 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.1 | 0.3 | GO:0048939 | anterior lateral line nerve glial cell differentiation(GO:0048913) myelination of anterior lateral line nerve axons(GO:0048914) anterior lateral line nerve glial cell development(GO:0048939) anterior lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048940) |
| 0.1 | 0.2 | GO:1990416 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.1 | 0.3 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 2.0 | GO:1903670 | regulation of sprouting angiogenesis(GO:1903670) |
| 0.1 | 0.5 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.1 | 0.2 | GO:1904088 | regulation of convergent extension involved in axis elongation(GO:1901232) positive regulation of epiboly involved in gastrulation with mouth forming second(GO:1904088) |
| 0.1 | 0.3 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.1 | 0.5 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.1 | 1.2 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.6 | GO:0014036 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.0 | 1.3 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.6 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.2 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.0 | 0.3 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.7 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.0 | 0.2 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.0 | 0.2 | GO:0071380 | response to prostaglandin E(GO:0034695) cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.0 | 0.1 | GO:0019418 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.0 | 0.8 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.3 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.0 | 0.6 | GO:0030947 | regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030947) |
| 0.0 | 0.3 | GO:0006537 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.0 | 2.7 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.2 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.0 | 0.2 | GO:0035889 | otolith tethering(GO:0035889) |
| 0.0 | 0.3 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.0 | 0.2 | GO:0003321 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) positive regulation of heart rate(GO:0010460) |
| 0.0 | 0.8 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.4 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.3 | GO:0030431 | sleep(GO:0030431) |
| 0.0 | 0.8 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.0 | 0.2 | GO:0006337 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.5 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.4 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.8 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.2 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.0 | 0.4 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.7 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.0 | 0.2 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.2 | GO:0045901 | positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 | 1.0 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.0 | 0.2 | GO:0031269 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.3 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.4 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.4 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.3 | GO:0033077 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.0 | 0.5 | GO:0003323 | glandular epithelial cell development(GO:0002068) type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.4 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.2 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.3 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.3 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.1 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 1.1 | GO:0014904 | myotube cell development(GO:0014904) skeletal muscle fiber development(GO:0048741) |
| 0.0 | 0.2 | GO:0046887 | positive regulation of hormone secretion(GO:0046887) |
| 0.0 | 0.1 | GO:1990118 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.0 | GO:0002828 | regulation of type 2 immune response(GO:0002828) |
| 0.0 | 0.4 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.2 | GO:0060396 | growth hormone receptor signaling pathway(GO:0060396) response to growth hormone(GO:0060416) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.4 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.1 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 0.1 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.0 | 0.5 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 0.2 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.5 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.1 | 0.4 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 0.6 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.1 | 0.3 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 1.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 0.7 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 0.9 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 1.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 0.3 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.1 | 0.3 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.5 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.2 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.0 | 0.7 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.2 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 1.0 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.4 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 2.0 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.2 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0030428 | cell septum(GO:0030428) |
| 0.0 | 0.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.1 | GO:0071914 | prominosome(GO:0071914) |
| 0.0 | 0.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.2 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.6 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 2.5 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.8 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.5 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.4 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 4.3 | GO:0031012 | extracellular matrix(GO:0031012) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.8 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.2 | 0.9 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.2 | 1.2 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.2 | 0.9 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.2 | 0.5 | GO:0031704 | apelin receptor binding(GO:0031704) |
| 0.1 | 4.1 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 0.4 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.1 | 0.4 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.1 | 0.5 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 0.5 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 0.7 | GO:0043121 | neurotrophin binding(GO:0043121) |
| 0.1 | 0.4 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.1 | 0.2 | GO:0001635 | adrenomedullin receptor activity(GO:0001605) calcitonin gene-related peptide receptor activity(GO:0001635) |
| 0.1 | 0.9 | GO:0043028 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.1 | 0.3 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.1 | 0.3 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.1 | 0.4 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 0.6 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 0.4 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 0.6 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 0.3 | GO:0038132 | neuregulin receptor activity(GO:0038131) neuregulin binding(GO:0038132) |
| 0.1 | 0.5 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.4 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.3 | GO:0031705 | bombesin receptor binding(GO:0031705) |
| 0.0 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.3 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.0 | 0.3 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.0 | 0.2 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.0 | 0.1 | GO:0070224 | sulfide:quinone oxidoreductase activity(GO:0070224) |
| 0.0 | 0.2 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.8 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 4.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 1.2 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.2 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 1.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.3 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.2 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.0 | 1.0 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.3 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.6 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.2 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.2 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.2 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.3 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 1.6 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.2 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 0.3 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.1 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.0 | 0.8 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.7 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.4 | GO:0008308 | voltage-gated chloride channel activity(GO:0005247) voltage-gated anion channel activity(GO:0008308) |
| 0.0 | 0.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.2 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.2 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.7 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.2 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.0 | 0.1 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.0 | 0.1 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.6 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.1 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.0 | 2.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 1.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.3 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.1 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.0 | 0.1 | GO:0015386 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.2 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 1.0 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.9 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.6 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.8 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 1.0 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.4 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.3 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.5 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 1.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 1.0 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 1.1 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.3 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.6 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.9 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.5 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.0 | 0.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.3 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.2 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |