Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
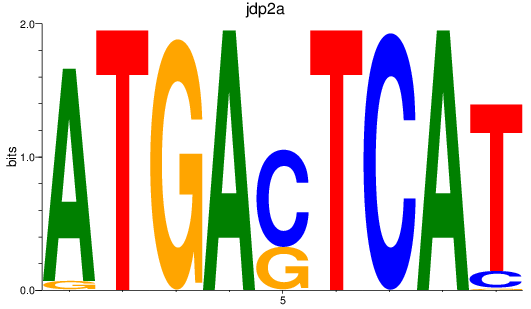
Results for jdp2a
Z-value: 1.00
Transcription factors associated with jdp2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
jdp2a
|
ENSDARG00000040137 | Jun dimerization protein 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| jdp2a | dr11_v1_chr17_-_50220228_50220228 | 0.58 | 1.0e-01 | Click! |
Activity profile of jdp2a motif
Sorted Z-values of jdp2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_44802353 | 1.16 |
ENSDART00000066380
|
ca7
|
carbonic anhydrase VII |
| chr8_-_34402839 | 0.97 |
ENSDART00000180173
ENSDART00000191588 ENSDART00000186376 |
gapvd1
|
GTPase activating protein and VPS9 domains 1 |
| chr22_+_23359369 | 0.94 |
ENSDART00000170886
|
dennd1b
|
DENN/MADD domain containing 1B |
| chr8_-_38201415 | 0.92 |
ENSDART00000155189
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr20_-_16548912 | 0.90 |
ENSDART00000137601
|
ches1
|
checkpoint suppressor 1 |
| chr8_-_22288004 | 0.83 |
ENSDART00000100042
|
si:ch211-147a11.3
|
si:ch211-147a11.3 |
| chr8_-_22288258 | 0.83 |
ENSDART00000140978
ENSDART00000100046 |
si:ch211-147a11.3
|
si:ch211-147a11.3 |
| chr15_+_25489406 | 0.76 |
ENSDART00000162482
|
zgc:152863
|
zgc:152863 |
| chr21_+_13233377 | 0.75 |
ENSDART00000142569
|
specc1lb
|
sperm antigen with calponin homology and coiled-coil domains 1-like b |
| chr2_+_15100742 | 0.74 |
ENSDART00000027171
|
f3b
|
coagulation factor IIIb |
| chr19_+_7001170 | 0.71 |
ENSDART00000110366
|
zbtb22b
|
zinc finger and BTB domain containing 22b |
| chr14_+_30774032 | 0.71 |
ENSDART00000139552
|
atl3
|
atlastin 3 |
| chr2_-_7605121 | 0.66 |
ENSDART00000182099
|
CABZ01021599.1
|
|
| chr20_-_35512932 | 0.62 |
ENSDART00000137690
|
adgrf3b
|
adhesion G protein-coupled receptor F3b |
| chr22_-_23267893 | 0.61 |
ENSDART00000105613
|
si:dkey-121a9.3
|
si:dkey-121a9.3 |
| chr5_+_52706316 | 0.60 |
ENSDART00000169488
|
apba1a
|
amyloid beta (A4) precursor protein-binding, family A, member 1a |
| chr21_+_3093419 | 0.59 |
ENSDART00000162520
|
SHC3
|
SHC adaptor protein 3 |
| chr19_-_5805923 | 0.59 |
ENSDART00000134340
|
si:ch211-264f5.8
|
si:ch211-264f5.8 |
| chr8_-_14375890 | 0.59 |
ENSDART00000090306
|
xpr1a
|
xenotropic and polytropic retrovirus receptor 1a |
| chr4_+_68087932 | 0.58 |
ENSDART00000150494
|
znf1096
|
zinc finger protein 1096 |
| chr9_-_22057658 | 0.57 |
ENSDART00000101944
|
crygmxl1
|
crystallin, gamma MX, like 1 |
| chr6_+_40354424 | 0.56 |
ENSDART00000047416
|
slc4a8
|
solute carrier family 4, sodium bicarbonate cotransporter, member 8 |
| chr12_-_10315039 | 0.54 |
ENSDART00000152680
|
pyyb
|
peptide YYb |
| chr4_+_45274792 | 0.53 |
ENSDART00000150295
|
znf1138
|
zinc finger protein 1138 |
| chr9_+_33334501 | 0.53 |
ENSDART00000006867
|
ddx3a
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3a |
| chr3_+_16976095 | 0.53 |
ENSDART00000112450
|
cavin1a
|
caveolae associated protein 1a |
| chr5_+_20035284 | 0.53 |
ENSDART00000191808
|
sgsm1a
|
small G protein signaling modulator 1a |
| chr4_+_64921810 | 0.52 |
ENSDART00000157654
|
BX928743.1
|
|
| chr6_+_59832786 | 0.51 |
ENSDART00000154985
ENSDART00000102148 |
ddx3b
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3b |
| chr12_-_6880694 | 0.51 |
ENSDART00000171846
|
pcdh15b
|
protocadherin-related 15b |
| chr2_-_42827336 | 0.51 |
ENSDART00000140913
|
adcy8
|
adenylate cyclase 8 (brain) |
| chr5_+_34549845 | 0.51 |
ENSDART00000139317
|
aif1l
|
allograft inflammatory factor 1-like |
| chr8_+_48966165 | 0.51 |
ENSDART00000165425
|
aak1a
|
AP2 associated kinase 1a |
| chr10_+_5954787 | 0.51 |
ENSDART00000161887
ENSDART00000160345 ENSDART00000190046 |
map3k1
|
mitogen-activated protein kinase kinase kinase 1, E3 ubiquitin protein ligase |
| chr16_-_24832038 | 0.50 |
ENSDART00000153731
|
si:dkey-79d12.5
|
si:dkey-79d12.5 |
| chr14_+_30774515 | 0.50 |
ENSDART00000191666
|
atl3
|
atlastin 3 |
| chr3_-_45250924 | 0.50 |
ENSDART00000109017
|
usp31
|
ubiquitin specific peptidase 31 |
| chr2_+_30379650 | 0.50 |
ENSDART00000129542
|
crispld1b
|
cysteine-rich secretory protein LCCL domain containing 1b |
| chr25_+_30196039 | 0.50 |
ENSDART00000005299
|
hsd17b12a
|
hydroxysteroid (17-beta) dehydrogenase 12a |
| chr24_-_5691956 | 0.49 |
ENSDART00000189112
|
dia1b
|
deleted in autism 1b |
| chr6_+_44197099 | 0.48 |
ENSDART00000124168
|
ppp4r2b
|
protein phosphatase 4, regulatory subunit 2b |
| chr14_+_30774894 | 0.48 |
ENSDART00000023054
|
atl3
|
atlastin 3 |
| chr3_+_35498119 | 0.48 |
ENSDART00000178963
|
tnrc6a
|
trinucleotide repeat containing 6a |
| chr6_-_43028896 | 0.48 |
ENSDART00000149977
|
glyctk
|
glycerate kinase |
| chr21_-_2248239 | 0.48 |
ENSDART00000169448
|
si:ch73-299h12.1
|
si:ch73-299h12.1 |
| chr5_-_54482245 | 0.48 |
ENSDART00000177470
|
fbxw5
|
F-box and WD repeat domain containing 5 |
| chr17_+_23556764 | 0.48 |
ENSDART00000146787
|
pank1a
|
pantothenate kinase 1a |
| chr8_+_48965767 | 0.47 |
ENSDART00000008058
|
aak1a
|
AP2 associated kinase 1a |
| chr6_+_44197348 | 0.47 |
ENSDART00000075486
|
ppp4r2b
|
protein phosphatase 4, regulatory subunit 2b |
| chr2_-_51087077 | 0.47 |
ENSDART00000167987
|
ftr67
|
finTRIM family, member 67 |
| chr5_-_64355227 | 0.47 |
ENSDART00000170787
|
fam78aa
|
family with sequence similarity 78, member Aa |
| chr9_+_33154841 | 0.47 |
ENSDART00000132465
|
dopey2
|
dopey family member 2 |
| chr3_+_53352018 | 0.46 |
ENSDART00000082715
|
camsap3
|
calmodulin regulated spectrin-associated protein family, member 3 |
| chr14_+_3507326 | 0.46 |
ENSDART00000159326
|
gstp1
|
glutathione S-transferase pi 1 |
| chr12_+_29173523 | 0.46 |
ENSDART00000153212
|
adgra1a
|
adhesion G protein-coupled receptor A1a |
| chr18_+_44849809 | 0.45 |
ENSDART00000097328
|
arfgap2
|
ADP-ribosylation factor GTPase activating protein 2 |
| chr25_-_13408760 | 0.44 |
ENSDART00000154445
|
gins3
|
GINS complex subunit 3 |
| chr7_-_60351876 | 0.44 |
ENSDART00000098563
|
plcb3
|
phospholipase C, beta 3 (phosphatidylinositol-specific) |
| chr23_+_39462208 | 0.44 |
ENSDART00000136707
|
mical1
|
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr2_-_32738535 | 0.43 |
ENSDART00000135293
|
nrbp2a
|
nuclear receptor binding protein 2a |
| chr23_+_39461989 | 0.42 |
ENSDART00000184761
|
mical1
|
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr13_+_7575563 | 0.42 |
ENSDART00000146592
|
gbf1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
| chr1_+_51312752 | 0.42 |
ENSDART00000063938
|
mast1a
|
microtubule associated serine/threonine kinase 1a |
| chr7_+_34549198 | 0.42 |
ENSDART00000173784
|
fhod1
|
formin homology 2 domain containing 1 |
| chr5_-_69523816 | 0.42 |
ENSDART00000112692
|
si:ch211-157p22.10
|
si:ch211-157p22.10 |
| chr5_+_65087226 | 0.42 |
ENSDART00000183187
|
ptrh1
|
peptidyl-tRNA hydrolase 1 homolog |
| chr5_-_20123002 | 0.41 |
ENSDART00000026516
|
pxmp2
|
peroxisomal membrane protein 2 |
| chr7_+_34549377 | 0.41 |
ENSDART00000191814
|
fhod1
|
formin homology 2 domain containing 1 |
| chr4_-_77267787 | 0.41 |
ENSDART00000190346
|
zgc:174310
|
zgc:174310 |
| chr4_-_64605318 | 0.41 |
ENSDART00000170391
|
znf1099
|
zinc finger protein 1099 |
| chr15_+_22867174 | 0.40 |
ENSDART00000035812
|
grik4
|
glutamate receptor, ionotropic, kainate 4 |
| chr7_-_5396154 | 0.38 |
ENSDART00000172980
|
arhgef11
|
Rho guanine nucleotide exchange factor (GEF) 11 |
| chr17_+_12942021 | 0.38 |
ENSDART00000192514
|
nfkbiab
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha b |
| chr15_+_29709382 | 0.38 |
ENSDART00000099956
|
nrip1a
|
nuclear receptor interacting protein 1a |
| chr13_-_7031033 | 0.38 |
ENSDART00000193211
|
CABZ01061524.1
|
|
| chr7_-_60351537 | 0.37 |
ENSDART00000159875
|
plcb3
|
phospholipase C, beta 3 (phosphatidylinositol-specific) |
| chr11_+_19370717 | 0.36 |
ENSDART00000165906
|
prickle2b
|
prickle homolog 2b |
| chr24_-_38192003 | 0.36 |
ENSDART00000109975
|
crp7
|
C-reactive protein 7 |
| chr18_+_14342326 | 0.36 |
ENSDART00000181013
ENSDART00000138372 |
si:dkey-246g23.2
|
si:dkey-246g23.2 |
| chr5_+_57480014 | 0.36 |
ENSDART00000135520
|
si:ch211-202f5.3
|
si:ch211-202f5.3 |
| chr4_-_64604842 | 0.36 |
ENSDART00000169862
|
znf1099
|
zinc finger protein 1099 |
| chr7_+_38770167 | 0.36 |
ENSDART00000190827
|
arhgap1
|
Rho GTPase activating protein 1 |
| chr4_-_72643171 | 0.35 |
ENSDART00000130126
|
CABZ01054394.1
|
|
| chr2_-_22966076 | 0.35 |
ENSDART00000143412
ENSDART00000146014 ENSDART00000183443 ENSDART00000191056 ENSDART00000183539 |
sap130b
|
Sin3A-associated protein b |
| chr18_-_41160579 | 0.35 |
ENSDART00000181480
|
CABZ01005876.1
|
|
| chr4_+_5317483 | 0.35 |
ENSDART00000150366
|
si:ch211-214j24.10
|
si:ch211-214j24.10 |
| chr6_+_49901465 | 0.35 |
ENSDART00000023515
|
chmp4ba
|
charged multivesicular body protein 4Ba |
| chr9_+_28140089 | 0.34 |
ENSDART00000046880
|
plekhm3
|
pleckstrin homology domain containing, family M, member 3 |
| chr3_+_19687217 | 0.34 |
ENSDART00000141937
|
tlk2
|
tousled-like kinase 2 |
| chr17_+_28675120 | 0.34 |
ENSDART00000159067
|
hectd1
|
HECT domain containing 1 |
| chr1_+_46598502 | 0.34 |
ENSDART00000132861
|
cab39l
|
calcium binding protein 39-like |
| chr16_+_27349585 | 0.34 |
ENSDART00000142573
|
nr4a3
|
nuclear receptor subfamily 4, group A, member 3 |
| chr12_+_23371542 | 0.34 |
ENSDART00000078896
|
waca
|
WW domain containing adaptor with coiled-coil a |
| chr12_-_35386910 | 0.34 |
ENSDART00000153453
|
camk2g1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II gamma 1 |
| chr8_-_14067517 | 0.33 |
ENSDART00000140948
|
dedd
|
death effector domain containing |
| chr11_+_24251141 | 0.33 |
ENSDART00000182684
|
pnp4a
|
purine nucleoside phosphorylase 4a |
| chr21_+_37477001 | 0.33 |
ENSDART00000114778
|
amot
|
angiomotin |
| chr22_-_5839686 | 0.33 |
ENSDART00000145406
|
si:rp71-36a1.5
|
si:rp71-36a1.5 |
| chr7_+_14005111 | 0.32 |
ENSDART00000187365
|
furina
|
furin (paired basic amino acid cleaving enzyme) a |
| chr6_-_33685325 | 0.32 |
ENSDART00000181883
|
mast2
|
microtubule associated serine/threonine kinase 2 |
| chr3_+_33440615 | 0.32 |
ENSDART00000146005
|
gtpbp1
|
GTP binding protein 1 |
| chr5_+_65086668 | 0.32 |
ENSDART00000183746
|
ptrh1
|
peptidyl-tRNA hydrolase 1 homolog |
| chr11_-_39118882 | 0.32 |
ENSDART00000113185
ENSDART00000156526 |
ap5b1
|
adaptor-related protein complex 5, beta 1 subunit |
| chr18_+_23218980 | 0.32 |
ENSDART00000185014
|
mef2aa
|
myocyte enhancer factor 2aa |
| chr18_-_19095141 | 0.32 |
ENSDART00000090962
ENSDART00000090922 |
dennd4a
|
DENN/MADD domain containing 4A |
| chr12_-_7639120 | 0.31 |
ENSDART00000126712
ENSDART00000126219 |
ccdc6b
|
coiled-coil domain containing 6b |
| chr19_-_21832441 | 0.31 |
ENSDART00000151272
ENSDART00000151442 ENSDART00000150168 ENSDART00000148797 ENSDART00000128196 ENSDART00000149259 ENSDART00000052556 ENSDART00000149658 ENSDART00000149639 ENSDART00000148424 |
mbpa
|
myelin basic protein a |
| chr1_-_11689525 | 0.31 |
ENSDART00000090689
|
brat1
|
BRCA1-associated ATM activator 1 |
| chr2_-_48356787 | 0.31 |
ENSDART00000098004
|
per2
|
period circadian clock 2 |
| chr14_+_20156477 | 0.31 |
ENSDART00000123434
|
fmr1
|
fragile X mental retardation 1 |
| chr3_+_19216567 | 0.30 |
ENSDART00000134433
|
il12rb2l
|
interleukin 12 receptor, beta 2a, like |
| chr4_-_38421217 | 0.30 |
ENSDART00000164517
|
znf1092
|
szinc finger protein 1092 |
| chr21_-_2173300 | 0.30 |
ENSDART00000172276
|
zgc:163077
|
zgc:163077 |
| chr18_+_16750080 | 0.30 |
ENSDART00000136320
|
rnf141
|
ring finger protein 141 |
| chr6_+_11250316 | 0.29 |
ENSDART00000137122
|
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr19_+_4139065 | 0.29 |
ENSDART00000172524
|
si:dkey-218f9.10
|
si:dkey-218f9.10 |
| chr24_-_31090948 | 0.29 |
ENSDART00000176799
|
hccsb
|
holocytochrome c synthase b |
| chr17_+_21486047 | 0.29 |
ENSDART00000104608
|
pla2g4f.2
|
phospholipase A2, group IVF, tandem duplicate 2 |
| chr14_-_493029 | 0.29 |
ENSDART00000115093
|
zgc:172215
|
zgc:172215 |
| chr19_-_31035325 | 0.29 |
ENSDART00000147504
|
bzw2
|
basic leucine zipper and W2 domains 2 |
| chr21_+_40498628 | 0.29 |
ENSDART00000163454
|
coro6
|
coronin 6 |
| chr15_-_30450898 | 0.29 |
ENSDART00000156584
|
msi2b
|
musashi RNA-binding protein 2b |
| chr5_+_65086856 | 0.29 |
ENSDART00000169209
ENSDART00000162409 |
ptrh1
|
peptidyl-tRNA hydrolase 1 homolog |
| chr13_+_23988442 | 0.28 |
ENSDART00000010918
|
agt
|
angiotensinogen |
| chr15_+_24704798 | 0.28 |
ENSDART00000192470
|
LRRC75A
|
si:dkey-151p21.7 |
| chr11_-_24458786 | 0.28 |
ENSDART00000089713
|
mxra8a
|
matrix-remodelling associated 8a |
| chr18_-_41160975 | 0.28 |
ENSDART00000187517
|
CABZ01005876.1
|
|
| chr21_-_2172924 | 0.27 |
ENSDART00000169262
|
zgc:163077
|
zgc:163077 |
| chr22_+_4442473 | 0.27 |
ENSDART00000170751
|
ticam1
|
toll-like receptor adaptor molecule 1 |
| chr13_+_30421472 | 0.27 |
ENSDART00000143569
|
zmiz1a
|
zinc finger, MIZ-type containing 1a |
| chr24_-_24797455 | 0.27 |
ENSDART00000138741
|
pde7a
|
phosphodiesterase 7A |
| chr3_+_17616201 | 0.27 |
ENSDART00000156775
|
rab5c
|
RAB5C, member RAS oncogene family |
| chr2_-_23391266 | 0.26 |
ENSDART00000159048
|
ivns1abpb
|
influenza virus NS1A binding protein b |
| chr2_-_37277626 | 0.26 |
ENSDART00000135340
|
nadkb
|
NAD kinase b |
| chr4_-_69550757 | 0.26 |
ENSDART00000168344
|
znf1101
|
zinc finger protein 1101 |
| chr7_+_528593 | 0.26 |
ENSDART00000091955
|
nrxn2b
|
neurexin 2b |
| chr8_+_24854600 | 0.26 |
ENSDART00000156570
|
slc6a17
|
solute carrier family 6 (neutral amino acid transporter), member 17 |
| chr5_+_59534286 | 0.25 |
ENSDART00000189341
|
gtf2ird1
|
GTF2I repeat domain containing 1 |
| chr6_+_42475730 | 0.25 |
ENSDART00000150226
|
mst1ra
|
macrophage stimulating 1 receptor a |
| chr6_-_12588044 | 0.25 |
ENSDART00000047896
|
slc15a1b
|
solute carrier family 15 (oligopeptide transporter), member 1b |
| chr4_+_58057168 | 0.24 |
ENSDART00000161097
|
si:dkey-57k17.1
|
si:dkey-57k17.1 |
| chr5_+_63767376 | 0.24 |
ENSDART00000138898
|
rgs3b
|
regulator of G protein signaling 3b |
| chr25_-_29072162 | 0.24 |
ENSDART00000169269
|
arid3b
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr16_-_30864824 | 0.24 |
ENSDART00000190444
ENSDART00000192231 |
ptk2ab
|
protein tyrosine kinase 2ab |
| chr19_-_31035155 | 0.24 |
ENSDART00000161882
|
bzw2
|
basic leucine zipper and W2 domains 2 |
| chr1_+_52137528 | 0.24 |
ENSDART00000007079
ENSDART00000074265 |
asna1
|
arsA arsenite transporter, ATP-binding, homolog 1 (bacterial) |
| chr7_+_6317866 | 0.24 |
ENSDART00000173397
|
si:ch211-220f21.3
|
si:ch211-220f21.3 |
| chr20_-_38746889 | 0.23 |
ENSDART00000140275
|
trim54
|
tripartite motif containing 54 |
| chr21_-_18824434 | 0.23 |
ENSDART00000156333
|
si:dkey-112m2.1
|
si:dkey-112m2.1 |
| chr1_-_54765262 | 0.23 |
ENSDART00000150362
|
si:ch211-197k17.3
|
si:ch211-197k17.3 |
| chr6_-_37744430 | 0.22 |
ENSDART00000150177
ENSDART00000149722 |
nipa2
|
non imprinted in Prader-Willi/Angelman syndrome 2 (human) |
| chr10_-_32877348 | 0.22 |
ENSDART00000018977
ENSDART00000133421 |
rabgef1
|
RAB guanine nucleotide exchange factor (GEF) 1 |
| chr11_+_19370447 | 0.21 |
ENSDART00000186154
|
prickle2b
|
prickle homolog 2b |
| chr11_-_25461336 | 0.20 |
ENSDART00000014945
|
hcfc1a
|
host cell factor C1a |
| chr4_-_56157199 | 0.20 |
ENSDART00000169806
|
si:ch211-207e19.15
|
si:ch211-207e19.15 |
| chr7_-_19369002 | 0.20 |
ENSDART00000165680
|
ntn4
|
netrin 4 |
| chr16_+_5908483 | 0.20 |
ENSDART00000167393
|
ulk4
|
unc-51 like kinase 4 |
| chr17_-_6536466 | 0.20 |
ENSDART00000188735
|
cenpo
|
centromere protein O |
| chr18_+_13077800 | 0.20 |
ENSDART00000161153
|
GAN
|
gigaxonin |
| chr24_-_21973365 | 0.19 |
ENSDART00000081204
ENSDART00000030592 |
acot9.1
|
acyl-CoA thioesterase 9, tandem duplicate 1 |
| chr19_+_26072624 | 0.19 |
ENSDART00000147627
|
jarid2b
|
jumonji, AT rich interactive domain 2b |
| chr1_+_46598764 | 0.19 |
ENSDART00000053240
|
cab39l
|
calcium binding protein 39-like |
| chr15_+_20352123 | 0.18 |
ENSDART00000011030
ENSDART00000163532 ENSDART00000169537 ENSDART00000161047 |
il15l
|
interleukin 15, like |
| chr12_+_18718065 | 0.18 |
ENSDART00000152935
|
mkl1b
|
megakaryoblastic leukemia (translocation) 1b |
| chr16_+_18974064 | 0.18 |
ENSDART00000079248
|
slc6a19b
|
solute carrier family 6 (neutral amino acid transporter), member 19b |
| chr16_+_46410520 | 0.18 |
ENSDART00000131072
|
rpz2
|
rapunzel 2 |
| chr5_+_50953240 | 0.17 |
ENSDART00000148501
ENSDART00000149892 ENSDART00000190312 |
col4a3bpa
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein a |
| chr5_-_63644938 | 0.16 |
ENSDART00000050865
|
surf4l
|
surfeit gene 4, like |
| chr23_+_4253957 | 0.16 |
ENSDART00000008761
|
arl6ip5b
|
ADP-ribosylation factor-like 6 interacting protein 5b |
| chr12_+_48390715 | 0.16 |
ENSDART00000149351
|
scd
|
stearoyl-CoA desaturase (delta-9-desaturase) |
| chr2_+_9822319 | 0.16 |
ENSDART00000144078
ENSDART00000144371 |
anxa13l
|
annexin A13, like |
| chr5_-_57480660 | 0.16 |
ENSDART00000147875
ENSDART00000142776 |
si:ch211-202f5.2
|
si:ch211-202f5.2 |
| chr22_-_16416882 | 0.16 |
ENSDART00000062749
|
cts12
|
cathepsin 12 |
| chr1_+_58602506 | 0.15 |
ENSDART00000158425
|
si:ch73-221f6.1
|
si:ch73-221f6.1 |
| chr6_-_43449013 | 0.15 |
ENSDART00000122423
|
eevs
|
2-epi-5-epi-valiolone synthase |
| chr4_-_45146463 | 0.14 |
ENSDART00000187483
|
znf1046
|
zinc finger protein 1046 |
| chr21_-_11657043 | 0.14 |
ENSDART00000141297
|
cast
|
calpastatin |
| chr14_+_24934736 | 0.14 |
ENSDART00000191821
|
ppargc1b
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta |
| chr20_+_28364742 | 0.14 |
ENSDART00000103355
|
rhov
|
ras homolog family member V |
| chr24_-_21973163 | 0.13 |
ENSDART00000131406
|
acot9.1
|
acyl-CoA thioesterase 9, tandem duplicate 1 |
| chr11_+_33312601 | 0.13 |
ENSDART00000188024
|
cntnap5l
|
contactin associated protein-like 5 like |
| chr3_+_24986145 | 0.13 |
ENSDART00000055428
|
cbx7a
|
chromobox homolog 7a |
| chr7_+_7019911 | 0.13 |
ENSDART00000172421
|
rbm14b
|
RNA binding motif protein 14b |
| chr21_+_26612777 | 0.13 |
ENSDART00000142667
|
esrra
|
estrogen-related receptor alpha |
| chr10_+_28428222 | 0.13 |
ENSDART00000135003
|
si:ch211-222e20.4
|
si:ch211-222e20.4 |
| chr5_+_24287927 | 0.13 |
ENSDART00000143563
|
zdhhc23a
|
zinc finger, DHHC-type containing 23a |
| chr11_-_25324534 | 0.13 |
ENSDART00000158598
|
si:ch211-232b12.5
|
si:ch211-232b12.5 |
| chr10_-_74408 | 0.12 |
ENSDART00000100073
ENSDART00000141723 |
dyrk1aa
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A, a |
| chr2_-_15362923 | 0.12 |
ENSDART00000166926
|
olfm3b
|
olfactomedin 3b |
| chr7_+_22688781 | 0.11 |
ENSDART00000173509
|
ugt5g1
|
UDP glucuronosyltransferase 5 family, polypeptide G1 |
| chr25_-_7753207 | 0.11 |
ENSDART00000126499
|
phf21ab
|
PHD finger protein 21Ab |
| chr17_-_6536305 | 0.11 |
ENSDART00000154855
|
cenpo
|
centromere protein O |
| chr20_+_23960525 | 0.11 |
ENSDART00000042123
|
cx52.6
|
connexin 52.6 |
| chr1_-_43905252 | 0.10 |
ENSDART00000135477
ENSDART00000132089 |
si:dkey-22i16.3
|
si:dkey-22i16.3 |
| chr17_+_33495194 | 0.10 |
ENSDART00000033691
|
pth2
|
parathyroid hormone 2 |
| chr1_+_6640437 | 0.10 |
ENSDART00000147638
|
si:ch211-93g23.2
|
si:ch211-93g23.2 |
| chr12_+_1455147 | 0.10 |
ENSDART00000018752
|
cops3
|
COP9 signalosome subunit 3 |
| chr19_+_33701366 | 0.10 |
ENSDART00000162517
|
runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr3_-_16719244 | 0.10 |
ENSDART00000055859
|
pold1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr9_-_30346279 | 0.09 |
ENSDART00000089488
|
sytl5
|
synaptotagmin-like 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of jdp2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.2 | 0.7 | GO:0097095 | frontonasal suture morphogenesis(GO:0097095) |
| 0.2 | 0.5 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.1 | 0.3 | GO:0035666 | regulation of natural killer cell activation(GO:0032814) positive regulation of natural killer cell activation(GO:0032816) TRIF-dependent toll-like receptor signaling pathway(GO:0035666) |
| 0.1 | 0.3 | GO:0015824 | proline transport(GO:0015824) |
| 0.1 | 0.3 | GO:0032615 | interleukin-12 production(GO:0032615) regulation of interleukin-12 production(GO:0032655) |
| 0.1 | 0.5 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.1 | 0.2 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 | 0.4 | GO:1902975 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.1 | 0.5 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 0.3 | GO:0017006 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.1 | 1.0 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.1 | 0.3 | GO:0044783 | G1 DNA damage checkpoint(GO:0044783) |
| 0.1 | 0.3 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.1 | 0.3 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 0.8 | GO:0090279 | regulation of calcium ion import(GO:0090279) |
| 0.0 | 0.1 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.3 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.5 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.3 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.0 | 0.2 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.5 | GO:0031111 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.0 | 0.3 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.3 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.5 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.8 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.5 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 1.7 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.5 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.3 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.0 | 1.7 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.0 | 0.3 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.5 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.0 | 0.1 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.3 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.3 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.4 | GO:0033505 | floor plate formation(GO:0021508) floor plate morphogenesis(GO:0033505) |
| 0.0 | 0.2 | GO:0070309 | lens fiber cell development(GO:0070307) lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.3 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.9 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.1 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.6 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.0 | 0.5 | GO:0010824 | regulation of centrosome duplication(GO:0010824) |
| 0.0 | 0.3 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.3 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.3 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.0 | 0.6 | GO:0032402 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.0 | 0.2 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.0 | 0.1 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.3 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.5 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.2 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.2 | GO:0001990 | regulation of systemic arterial blood pressure by hormone(GO:0001990) |
| 0.0 | 0.4 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.3 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.1 | 0.4 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.1 | 0.4 | GO:0000811 | GINS complex(GO:0000811) |
| 0.1 | 0.3 | GO:0031511 | Mis6-Sim4 complex(GO:0031511) |
| 0.1 | 0.3 | GO:1902737 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.0 | 0.5 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 1.7 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.3 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.4 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.5 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.3 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.1 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 0.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.5 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.5 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.5 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.3 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.1 | 1.0 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 0.5 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.1 | 0.5 | GO:0008887 | glycerate kinase activity(GO:0008887) |
| 0.1 | 0.5 | GO:0031841 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.1 | 0.4 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 1.0 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 0.5 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.5 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.1 | 0.4 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 0.3 | GO:0004408 | holocytochrome-c synthase activity(GO:0004408) |
| 0.0 | 0.9 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.2 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.0 | 0.6 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.3 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.3 | GO:0071916 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.0 | 0.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.8 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.6 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.1 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.6 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.1 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.2 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.5 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.1 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.5 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.9 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.1 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.0 | 0.3 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.3 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.5 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.1 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.1 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.8 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.8 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.2 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.6 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 0.8 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.5 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.5 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.4 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.6 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.3 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.5 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.5 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 0.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.3 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |