Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
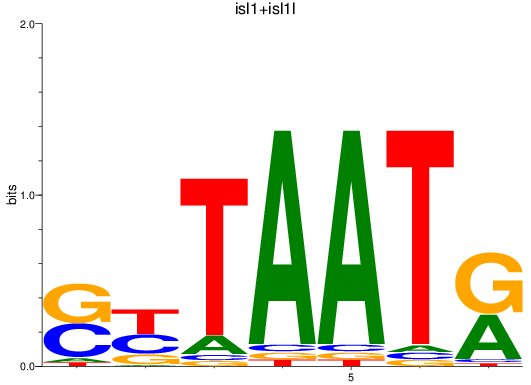
Results for isl1+isl1l
Z-value: 0.92
Transcription factors associated with isl1+isl1l
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
isl1
|
ENSDARG00000004023 | ISL LIM homeobox 1 |
|
isl1l
|
ENSDARG00000021055 | islet1, like |
|
isl1l
|
ENSDARG00000110204 | islet1, like |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| isl1 | dr11_v1_chr5_-_40734045_40734045 | 0.70 | 3.7e-02 | Click! |
| isl1l | dr11_v1_chr10_+_8680730_8680730 | 0.21 | 5.9e-01 | Click! |
Activity profile of isl1+isl1l motif
Sorted Z-values of isl1+isl1l motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_25679339 | 1.24 |
ENSDART00000161703
ENSDART00000054230 |
fgg
|
fibrinogen gamma chain |
| chr1_+_44439661 | 1.11 |
ENSDART00000100309
|
crybb1l2
|
crystallin, beta B1, like 2 |
| chr5_-_30615901 | 1.07 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr4_+_7508316 | 0.89 |
ENSDART00000170924
ENSDART00000170933 ENSDART00000164985 ENSDART00000167571 ENSDART00000158843 ENSDART00000158999 |
tnnt2e
|
troponin T2e, cardiac |
| chr23_+_36653376 | 0.87 |
ENSDART00000053189
|
gpr182
|
G protein-coupled receptor 182 |
| chr19_+_10855158 | 0.85 |
ENSDART00000172219
ENSDART00000170826 |
apoea
|
apolipoprotein Ea |
| chr6_+_15268685 | 0.78 |
ENSDART00000128090
ENSDART00000154417 |
ecrg4b
|
esophageal cancer related gene 4b |
| chr21_+_28958471 | 0.77 |
ENSDART00000144331
ENSDART00000005929 |
ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr9_+_7548533 | 0.72 |
ENSDART00000081543
|
ptprna
|
protein tyrosine phosphatase, receptor type, Na |
| chr5_+_32345187 | 0.71 |
ENSDART00000147132
|
c9
|
complement component 9 |
| chr3_+_15505275 | 0.69 |
ENSDART00000141714
|
nupr1
|
nuclear protein 1 |
| chr13_-_36391496 | 0.67 |
ENSDART00000100217
ENSDART00000140243 |
actn1
|
actinin, alpha 1 |
| chr22_-_14115292 | 0.64 |
ENSDART00000105717
ENSDART00000165670 |
aox5
|
aldehyde oxidase 5 |
| chr14_-_17072736 | 0.63 |
ENSDART00000106333
|
phox2bb
|
paired-like homeobox 2bb |
| chr2_+_59527149 | 0.61 |
ENSDART00000168568
|
CU861477.1
|
|
| chr23_+_36063599 | 0.60 |
ENSDART00000103147
|
hoxc12a
|
homeobox C12a |
| chr4_+_76705830 | 0.58 |
ENSDART00000064312
|
ms4a17a.7
|
membrane-spanning 4-domains, subfamily A, member 17A.7 |
| chr14_-_17121676 | 0.58 |
ENSDART00000170154
ENSDART00000060479 |
smtnl1
|
smoothelin-like 1 |
| chr6_+_2093206 | 0.56 |
ENSDART00000114314
|
tgm2b
|
transglutaminase 2b |
| chr20_+_572037 | 0.56 |
ENSDART00000028062
ENSDART00000152736 ENSDART00000031759 ENSDART00000162198 |
smyd2b
|
SET and MYND domain containing 2b |
| chr18_+_15132112 | 0.56 |
ENSDART00000099764
|
zgc:153031
|
zgc:153031 |
| chr10_+_32104305 | 0.54 |
ENSDART00000099880
|
wnt11r
|
wingless-type MMTV integration site family, member 11, related |
| chr6_+_2093367 | 0.54 |
ENSDART00000148396
|
tgm2b
|
transglutaminase 2b |
| chr20_+_2281933 | 0.53 |
ENSDART00000137579
|
si:ch73-18b11.2
|
si:ch73-18b11.2 |
| chr21_-_35853245 | 0.53 |
ENSDART00000172245
|
sgcd
|
sarcoglycan, delta (dystrophin-associated glycoprotein) |
| chr25_-_27541621 | 0.52 |
ENSDART00000130678
|
spam1
|
sperm adhesion molecule 1 |
| chr8_-_1051438 | 0.51 |
ENSDART00000067093
ENSDART00000170737 |
smyd1b
|
SET and MYND domain containing 1b |
| chr17_+_32343121 | 0.51 |
ENSDART00000156051
|
dhx32b
|
DEAH (Asp-Glu-Ala-His) box polypeptide 32b |
| chr24_+_36636208 | 0.50 |
ENSDART00000139211
|
si:ch73-334d15.4
|
si:ch73-334d15.4 |
| chr4_+_15968483 | 0.50 |
ENSDART00000101575
|
si:dkey-117n7.5
|
si:dkey-117n7.5 |
| chr15_-_24869826 | 0.50 |
ENSDART00000127047
|
tusc5a
|
tumor suppressor candidate 5a |
| chr3_-_58650057 | 0.49 |
ENSDART00000057640
|
dhrs7ca
|
dehydrogenase/reductase (SDR family) member 7Ca |
| chr23_-_31372639 | 0.48 |
ENSDART00000179908
ENSDART00000135620 ENSDART00000053367 |
hmgn3
|
high mobility group nucleosomal binding domain 3 |
| chr8_+_23213320 | 0.46 |
ENSDART00000032996
ENSDART00000137536 |
ppdpfa
|
pancreatic progenitor cell differentiation and proliferation factor a |
| chr5_-_64203101 | 0.46 |
ENSDART00000029364
|
ak5l
|
adenylate kinase 5, like |
| chr7_+_8543317 | 0.45 |
ENSDART00000114998
|
jac8
|
jacalin 8 |
| chr22_-_10121880 | 0.45 |
ENSDART00000002348
|
rdh5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr2_+_27010439 | 0.45 |
ENSDART00000030547
|
cdh7a
|
cadherin 7a |
| chr23_+_23020709 | 0.45 |
ENSDART00000146463
|
samd11
|
sterile alpha motif domain containing 11 |
| chr10_+_21576909 | 0.44 |
ENSDART00000168604
ENSDART00000166533 |
pcdh1a3
|
protocadherin 1 alpha 3 |
| chr16_+_24626448 | 0.44 |
ENSDART00000153591
|
si:dkey-56f14.7
|
si:dkey-56f14.7 |
| chr14_+_11457500 | 0.44 |
ENSDART00000169202
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr6_-_21492752 | 0.44 |
ENSDART00000006843
ENSDART00000171479 |
cacng1a
|
calcium channel, voltage-dependent, gamma subunit 1a |
| chr24_-_32665283 | 0.42 |
ENSDART00000038364
|
ca2
|
carbonic anhydrase II |
| chr6_+_16031189 | 0.42 |
ENSDART00000015333
|
gbx2
|
gastrulation brain homeobox 2 |
| chr4_+_5506952 | 0.41 |
ENSDART00000032857
ENSDART00000160222 |
mapk11
|
mitogen-activated protein kinase 11 |
| chr5_-_41831646 | 0.41 |
ENSDART00000134326
|
si:dkey-65b12.6
|
si:dkey-65b12.6 |
| chr6_+_16406723 | 0.40 |
ENSDART00000040035
|
ccdc80l1
|
coiled-coil domain containing 80 like 1 |
| chr1_+_41588170 | 0.40 |
ENSDART00000139175
|
si:dkey-56e3.2
|
si:dkey-56e3.2 |
| chr16_+_32059785 | 0.39 |
ENSDART00000134459
|
si:dkey-40m6.8
|
si:dkey-40m6.8 |
| chr15_-_23529945 | 0.39 |
ENSDART00000152543
|
hmbsb
|
hydroxymethylbilane synthase, b |
| chr13_-_16222388 | 0.39 |
ENSDART00000182861
|
zgc:110045
|
zgc:110045 |
| chr19_+_9786722 | 0.38 |
ENSDART00000138310
|
cacng6a
|
calcium channel, voltage-dependent, gamma subunit 6a |
| chr8_+_18830759 | 0.38 |
ENSDART00000089079
|
mpnd
|
MPN domain containing |
| chr5_-_42872712 | 0.38 |
ENSDART00000003947
|
flot2a
|
flotillin 2a |
| chr17_-_19535328 | 0.38 |
ENSDART00000077809
|
cyp26c1
|
cytochrome P450, family 26, subfamily C, polypeptide 1 |
| chr10_+_21797276 | 0.38 |
ENSDART00000169105
|
pcdh1g29
|
protocadherin 1 gamma 29 |
| chr7_+_69841017 | 0.38 |
ENSDART00000169107
|
FO818704.1
|
|
| chr10_-_22845485 | 0.36 |
ENSDART00000079454
|
vamp2
|
vesicle-associated membrane protein 2 |
| chr5_+_26795773 | 0.36 |
ENSDART00000145631
|
tcn2
|
transcobalamin II |
| chr6_-_19023468 | 0.36 |
ENSDART00000184729
|
sept9b
|
septin 9b |
| chr11_+_2172335 | 0.35 |
ENSDART00000170593
|
hoxc12b
|
homeobox C12b |
| chr1_-_19079957 | 0.34 |
ENSDART00000141795
|
phox2ba
|
paired-like homeobox 2ba |
| chr7_+_36035432 | 0.34 |
ENSDART00000179004
|
irx3a
|
iroquois homeobox 3a |
| chr5_-_55600689 | 0.34 |
ENSDART00000013229
|
gnaq
|
guanine nucleotide binding protein (G protein), q polypeptide |
| chr14_-_1454045 | 0.34 |
ENSDART00000161460
|
pmt
|
phosphoethanolamine methyltransferase |
| chr3_-_58116314 | 0.34 |
ENSDART00000154901
|
si:ch211-256e16.6
|
si:ch211-256e16.6 |
| chr14_+_20893065 | 0.33 |
ENSDART00000079452
|
lygl1
|
lysozyme g-like 1 |
| chr19_+_14921000 | 0.33 |
ENSDART00000144052
|
oprd1a
|
opioid receptor, delta 1a |
| chr4_+_2230701 | 0.32 |
ENSDART00000080439
|
cmah
|
cytidine monophospho-N-acetylneuraminic acid hydroxylase |
| chr17_+_42274825 | 0.32 |
ENSDART00000020156
|
pax1a
|
paired box 1a |
| chr25_+_459901 | 0.32 |
ENSDART00000154895
|
prtgb
|
protogenin homolog b (Gallus gallus) |
| chr12_+_38556462 | 0.31 |
ENSDART00000021069
ENSDART00000145377 |
rpl38
|
ribosomal protein L38 |
| chr16_+_53203370 | 0.31 |
ENSDART00000154669
|
si:ch211-269k10.2
|
si:ch211-269k10.2 |
| chr20_-_22464250 | 0.31 |
ENSDART00000165904
|
pdgfra
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr9_+_18716485 | 0.30 |
ENSDART00000135125
|
serp2
|
stress-associated endoplasmic reticulum protein family member 2 |
| chr7_+_56098590 | 0.30 |
ENSDART00000098453
|
cdh15
|
cadherin 15, type 1, M-cadherin (myotubule) |
| chr4_+_15899668 | 0.30 |
ENSDART00000101592
|
col7a1l
|
collagen type VII alpha 1-like |
| chr4_-_24019711 | 0.29 |
ENSDART00000077926
|
celf2
|
cugbp, Elav-like family member 2 |
| chr9_+_14152211 | 0.29 |
ENSDART00000148055
|
si:ch211-67e16.11
|
si:ch211-67e16.11 |
| chr1_+_51475094 | 0.29 |
ENSDART00000146352
|
meis1a
|
Meis homeobox 1 a |
| chr11_-_1400507 | 0.28 |
ENSDART00000173029
ENSDART00000172953 ENSDART00000111140 |
rpl29
|
ribosomal protein L29 |
| chr1_-_43727012 | 0.28 |
ENSDART00000181064
|
bdh2
|
3-hydroxybutyrate dehydrogenase, type 2 |
| chr21_-_43606502 | 0.27 |
ENSDART00000151030
|
si:ch73-362m14.4
|
si:ch73-362m14.4 |
| chr9_-_4506819 | 0.27 |
ENSDART00000113975
|
kcnj3a
|
potassium inwardly-rectifying channel, subfamily J, member 3a |
| chr22_-_36875264 | 0.27 |
ENSDART00000137548
|
kng1
|
kininogen 1 |
| chr3_+_46457450 | 0.26 |
ENSDART00000127127
|
si:ch211-66e2.5
|
si:ch211-66e2.5 |
| chr16_-_22192006 | 0.26 |
ENSDART00000163338
|
il6r
|
interleukin 6 receptor |
| chr21_-_22317920 | 0.26 |
ENSDART00000191083
ENSDART00000108701 |
gdpd4b
|
glycerophosphodiester phosphodiesterase domain containing 4b |
| chr22_-_13851297 | 0.26 |
ENSDART00000080306
|
s100b
|
S100 calcium binding protein, beta (neural) |
| chr1_-_43727418 | 0.25 |
ENSDART00000133715
ENSDART00000074597 ENSDART00000132542 ENSDART00000181792 |
bdh2
SLC9B2
|
3-hydroxybutyrate dehydrogenase, type 2 si:dkey-162b23.4 |
| chr25_-_14398281 | 0.25 |
ENSDART00000046934
|
coq9
|
coenzyme Q9 homolog (S. cerevisiae) |
| chr8_-_6943155 | 0.24 |
ENSDART00000139545
ENSDART00000033294 |
wdr13
|
WD repeat domain 13 |
| chr15_-_39969988 | 0.24 |
ENSDART00000146054
|
rps5
|
ribosomal protein S5 |
| chr4_-_25485404 | 0.24 |
ENSDART00000041402
|
pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr6_-_27123327 | 0.23 |
ENSDART00000073881
|
agxta
|
alanine-glyoxylate aminotransferase a |
| chr7_-_6680622 | 0.23 |
ENSDART00000180437
ENSDART00000173270 ENSDART00000066450 |
dusp19b
|
dual specificity phosphatase 19b |
| chr16_+_52999778 | 0.23 |
ENSDART00000011506
|
nkd2a
|
naked cuticle homolog 2a |
| chr13_-_16191674 | 0.23 |
ENSDART00000147868
|
vwc2
|
von Willebrand factor C domain containing 2 |
| chr21_-_20381481 | 0.22 |
ENSDART00000115236
|
atp5mea
|
ATP synthase membrane subunit ea |
| chr9_-_19161982 | 0.22 |
ENSDART00000081878
|
pou1f1
|
POU class 1 homeobox 1 |
| chr7_+_36898850 | 0.22 |
ENSDART00000113342
|
tox3
|
TOX high mobility group box family member 3 |
| chr15_+_36309070 | 0.22 |
ENSDART00000157034
|
gmnc
|
geminin coiled-coil domain containing |
| chr25_+_19489370 | 0.22 |
ENSDART00000091132
|
glsl
|
glutaminase like |
| chr7_+_38897836 | 0.22 |
ENSDART00000024330
|
creb3l1
|
cAMP responsive element binding protein 3-like 1 |
| chr4_-_25485214 | 0.22 |
ENSDART00000159941
|
pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr11_+_16216909 | 0.21 |
ENSDART00000081035
ENSDART00000147190 |
slc25a26
|
solute carrier family 25 (S-adenosylmethionine carrier), member 26 |
| chr6_-_49898881 | 0.21 |
ENSDART00000150204
|
atp5f1e
|
ATP synthase F1 subunit epsilon |
| chr5_-_41838354 | 0.21 |
ENSDART00000146793
|
si:dkey-65b12.6
|
si:dkey-65b12.6 |
| chr3_+_22377312 | 0.20 |
ENSDART00000155597
|
arhgap27l
|
Rho GTPase activating protein 27, like |
| chr17_-_25397558 | 0.20 |
ENSDART00000182052
|
fam167b
|
family with sequence similarity 167, member B |
| chr4_-_69189894 | 0.20 |
ENSDART00000169596
|
si:ch211-209j12.1
|
si:ch211-209j12.1 |
| chr20_+_34770197 | 0.20 |
ENSDART00000018304
|
mcm3
|
minichromosome maintenance complex component 3 |
| chr10_+_38708099 | 0.20 |
ENSDART00000172306
|
tmprss2
|
transmembrane protease, serine 2 |
| chr1_+_50997156 | 0.20 |
ENSDART00000010449
|
ugp2a
|
UDP-glucose pyrophosphorylase 2a |
| chr6_-_436658 | 0.19 |
ENSDART00000191515
|
grap2b
|
GRB2-related adaptor protein 2b |
| chr17_-_38769781 | 0.19 |
ENSDART00000062067
|
dglucy
|
D-glutamate cyclase |
| chr18_-_14743659 | 0.19 |
ENSDART00000125979
|
tshz3a
|
teashirt zinc finger homeobox 3a |
| chr9_+_44430974 | 0.19 |
ENSDART00000056846
|
ppp1r1c
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr12_-_46959990 | 0.19 |
ENSDART00000084557
|
lhpp
|
phospholysine phosphohistidine inorganic pyrophosphate phosphatase |
| chr25_+_35913614 | 0.19 |
ENSDART00000022437
|
gpia
|
glucose-6-phosphate isomerase a |
| chr16_-_46619967 | 0.18 |
ENSDART00000158341
|
tmem176l.3a
|
transmembrane protein 176l.3a |
| chr20_+_32406011 | 0.18 |
ENSDART00000018640
ENSDART00000137910 |
snx3
|
sorting nexin 3 |
| chr7_+_53541173 | 0.18 |
ENSDART00000159449
|
gramd2aa
|
GRAM domain containing 2Aa |
| chr15_+_16387088 | 0.18 |
ENSDART00000101789
|
flot2b
|
flotillin 2b |
| chr8_-_49172418 | 0.17 |
ENSDART00000142437
ENSDART00000036157 |
fkbp1aa
|
FK506 binding protein 1Aa |
| chr15_-_1962326 | 0.17 |
ENSDART00000179669
|
dock10
|
dedicator of cytokinesis 10 |
| chr4_+_14826299 | 0.17 |
ENSDART00000067035
|
shisal1a
|
shisa like 1a |
| chr7_-_18598661 | 0.17 |
ENSDART00000182109
|
DTX4
|
si:ch211-119e14.2 |
| chr8_+_23009662 | 0.16 |
ENSDART00000030885
|
uckl1a
|
uridine-cytidine kinase 1-like 1a |
| chr20_-_39333657 | 0.16 |
ENSDART00000153720
ENSDART00000142164 |
ccl38.1
|
chemokine (C-C motif) ligand 38, duplicate 1 |
| chr22_-_26852516 | 0.16 |
ENSDART00000005829
|
gde1
|
glycerophosphodiester phosphodiesterase 1 |
| chr3_-_22212764 | 0.16 |
ENSDART00000155490
|
maptb
|
microtubule-associated protein tau b |
| chr7_+_15871156 | 0.16 |
ENSDART00000145946
|
pax6b
|
paired box 6b |
| chr17_-_46457622 | 0.16 |
ENSDART00000130215
|
TMEM179 (1 of many)
|
transmembrane protein 179 |
| chr4_+_17280868 | 0.16 |
ENSDART00000145349
|
bcat1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr10_-_13116337 | 0.16 |
ENSDART00000164568
|
musk
|
muscle, skeletal, receptor tyrosine kinase |
| chr21_-_39327223 | 0.15 |
ENSDART00000115097
|
aifm5
|
apoptosis-inducing factor, mitochondrion-associated, 5 |
| chr16_+_9762261 | 0.15 |
ENSDART00000020654
|
psmd4b
|
proteasome 26S subunit, non-ATPase 4b |
| chr11_+_41459408 | 0.15 |
ENSDART00000182285
|
park7
|
parkinson protein 7 |
| chr19_+_10331325 | 0.15 |
ENSDART00000143930
|
tmem238a
|
transmembrane protein 238a |
| chr7_+_756942 | 0.15 |
ENSDART00000152224
|
zgc:63470
|
zgc:63470 |
| chr18_-_40509006 | 0.15 |
ENSDART00000021372
|
chrna5
|
cholinergic receptor, nicotinic, alpha 5 |
| chr8_+_54055390 | 0.15 |
ENSDART00000102696
|
magi1a
|
membrane associated guanylate kinase, WW and PDZ domain containing 1a |
| chr4_-_5856200 | 0.14 |
ENSDART00000121936
|
slc5a8
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 8 |
| chr22_-_36770649 | 0.14 |
ENSDART00000169442
|
acy1
|
aminoacylase 1 |
| chr2_+_13056802 | 0.14 |
ENSDART00000142649
|
prkag2b
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit b |
| chr7_-_34192834 | 0.14 |
ENSDART00000125131
|
smad6a
|
SMAD family member 6a |
| chr24_+_744713 | 0.14 |
ENSDART00000067764
|
stk17a
|
serine/threonine kinase 17a |
| chr5_-_57528943 | 0.14 |
ENSDART00000130320
|
pisd
|
phosphatidylserine decarboxylase |
| chr21_+_723998 | 0.14 |
ENSDART00000160956
|
oaz1b
|
ornithine decarboxylase antizyme 1b |
| chr8_-_30338872 | 0.14 |
ENSDART00000137583
|
dock8
|
dedicator of cytokinesis 8 |
| chr16_+_14029283 | 0.13 |
ENSDART00000146165
ENSDART00000132075 |
rusc1
|
RUN and SH3 domain containing 1 |
| chr10_+_17875891 | 0.13 |
ENSDART00000191744
|
phf24
|
PHD finger protein 24 |
| chr14_-_36799280 | 0.13 |
ENSDART00000168615
|
rnf130
|
ring finger protein 130 |
| chr7_-_30174882 | 0.13 |
ENSDART00000110409
|
frmd5
|
FERM domain containing 5 |
| chr15_+_9053059 | 0.13 |
ENSDART00000012039
|
ppm1na
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Na (putative) |
| chr4_-_75707991 | 0.13 |
ENSDART00000166358
ENSDART00000160021 |
si:dkey-165o8.2
|
si:dkey-165o8.2 |
| chr5_-_42083363 | 0.13 |
ENSDART00000162596
|
cxcl11.5
|
chemokine (C-X-C motif) ligand 11, duplicate 5 |
| chr7_+_15871408 | 0.13 |
ENSDART00000014572
|
pax6b
|
paired box 6b |
| chr9_+_4250918 | 0.13 |
ENSDART00000164543
|
kalrna
|
kalirin RhoGEF kinase a |
| chr7_-_33949246 | 0.12 |
ENSDART00000173513
|
pias1a
|
protein inhibitor of activated STAT, 1a |
| chr23_+_4022620 | 0.12 |
ENSDART00000055099
|
b4galt5
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 |
| chr1_-_35924495 | 0.12 |
ENSDART00000184424
|
smad1
|
SMAD family member 1 |
| chr4_+_3980247 | 0.12 |
ENSDART00000049194
|
gpr37b
|
G protein-coupled receptor 37b |
| chr15_-_12011202 | 0.12 |
ENSDART00000160427
ENSDART00000168715 |
si:dkey-202l22.6
|
si:dkey-202l22.6 |
| chr10_-_17587832 | 0.12 |
ENSDART00000113101
|
smarcad1b
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 b |
| chr1_+_38776294 | 0.12 |
ENSDART00000170546
|
wdr17
|
WD repeat domain 17 |
| chr12_+_26706745 | 0.12 |
ENSDART00000141401
|
arhgap12b
|
Rho GTPase activating protein 12b |
| chr22_+_7486867 | 0.11 |
ENSDART00000034586
|
CELA1 (1 of many)
|
zgc:112302 |
| chr12_-_48006835 | 0.11 |
ENSDART00000108989
|
adamts14
|
ADAM metallopeptidase with thrombospondin type 1 motif, 14 |
| chr11_+_27347076 | 0.11 |
ENSDART00000173383
|
fbln2
|
fibulin 2 |
| chr20_+_2039518 | 0.11 |
ENSDART00000043157
|
CABZ01088134.1
|
|
| chr24_-_36802891 | 0.11 |
ENSDART00000088130
|
dcakd
|
dephospho-CoA kinase domain containing |
| chr23_+_309099 | 0.11 |
ENSDART00000134556
ENSDART00000055152 |
taf11
|
TAF11 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr6_-_35738836 | 0.11 |
ENSDART00000111642
|
brinp3a.1
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3a, tandem duplicate 1 |
| chr8_-_25033681 | 0.11 |
ENSDART00000003493
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr12_+_31735159 | 0.11 |
ENSDART00000185442
|
RNF157
|
si:dkey-49c17.3 |
| chr3_-_23474416 | 0.10 |
ENSDART00000161672
|
si:dkey-225f5.5
|
si:dkey-225f5.5 |
| chr13_+_44857087 | 0.10 |
ENSDART00000017770
|
zbtb8os
|
zinc finger and BTB domain containing 8 opposite strand |
| chr20_+_15982482 | 0.10 |
ENSDART00000020999
|
angptl1a
|
angiopoietin-like 1a |
| chr11_-_279328 | 0.10 |
ENSDART00000066179
|
npffl
|
neuropeptide FF-amide peptide precursor like |
| chr9_+_38624478 | 0.10 |
ENSDART00000077389
ENSDART00000114728 |
znf148
|
zinc finger protein 148 |
| chr14_-_2196267 | 0.10 |
ENSDART00000161674
ENSDART00000125674 |
pcdh2ab8
pcdh2ab9
|
protocadherin 2 alpha b 8 protocadherin 2 alpha b 9 |
| chr15_-_15983183 | 0.10 |
ENSDART00000154841
|
synrg
|
synergin, gamma |
| chr4_+_9478500 | 0.10 |
ENSDART00000030738
|
lmf2b
|
lipase maturation factor 2b |
| chr14_+_23709134 | 0.09 |
ENSDART00000191162
ENSDART00000179754 ENSDART00000054266 |
gnpda1
|
glucosamine-6-phosphate deaminase 1 |
| chr2_+_1881334 | 0.09 |
ENSDART00000161420
|
adgrl2b.1
|
adhesion G protein-coupled receptor L2b, tandem duplicate 1 |
| chr9_+_38684871 | 0.09 |
ENSDART00000147938
ENSDART00000004462 |
heg1
|
heart development protein with EGF-like domains 1 |
| chr13_+_27314795 | 0.09 |
ENSDART00000128726
|
eef1a1a
|
eukaryotic translation elongation factor 1 alpha 1a |
| chr3_-_11008532 | 0.09 |
ENSDART00000165086
|
CR382337.3
|
|
| chr3_-_17716322 | 0.09 |
ENSDART00000192664
|
CABZ01018956.1
|
|
| chr1_+_9994811 | 0.09 |
ENSDART00000143719
ENSDART00000110749 |
si:dkeyp-75b4.10
|
si:dkeyp-75b4.10 |
| chr15_+_1199407 | 0.09 |
ENSDART00000163827
|
mfsd1
|
major facilitator superfamily domain containing 1 |
| chr6_-_40581376 | 0.09 |
ENSDART00000185412
|
tspo
|
translocator protein |
| chr14_+_21222287 | 0.09 |
ENSDART00000159905
|
si:ch211-175m2.4
|
si:ch211-175m2.4 |
| chr6_-_18618106 | 0.08 |
ENSDART00000161562
|
znf207b
|
zinc finger protein 207, b |
| chr22_+_26853254 | 0.08 |
ENSDART00000182487
|
tmem186
|
transmembrane protein 186 |
| chr2_-_44280061 | 0.08 |
ENSDART00000136818
|
mpz
|
myelin protein zero |
| chr9_-_7640692 | 0.08 |
ENSDART00000135616
|
dnajb2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of isl1+isl1l
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.2 | 0.7 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.2 | 0.8 | GO:0051000 | regulation of nitric-oxide synthase activity(GO:0050999) positive regulation of nitric-oxide synthase activity(GO:0051000) beta-amyloid clearance(GO:0097242) regulation of beta-amyloid clearance(GO:1900221) |
| 0.2 | 0.8 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.2 | 0.5 | GO:0035773 | insulin secretion involved in cellular response to glucose stimulus(GO:0035773) regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.2 | 0.6 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.1 | 0.6 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.1 | 0.4 | GO:0006824 | cobalt ion transport(GO:0006824) cobalamin transport(GO:0015889) |
| 0.1 | 0.3 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 0.4 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.1 | 0.5 | GO:0019184 | nonribosomal peptide biosynthetic process(GO:0019184) |
| 0.1 | 1.2 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.1 | 0.3 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.1 | 0.6 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 0.4 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.1 | 1.1 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 0.4 | GO:0003151 | outflow tract morphogenesis(GO:0003151) atrial cardiac muscle cell development(GO:0055014) |
| 0.1 | 0.3 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.1 | 0.3 | GO:0003311 | pancreatic D cell differentiation(GO:0003311) pancreatic epsilon cell differentiation(GO:0090104) |
| 0.1 | 0.8 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.1 | 0.2 | GO:2000378 | negative regulation of reactive oxygen species metabolic process(GO:2000378) |
| 0.0 | 0.5 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.0 | 0.4 | GO:0021871 | forebrain regionalization(GO:0021871) |
| 0.0 | 0.3 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.0 | 0.5 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.2 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.0 | 0.4 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.5 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.2 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.0 | 0.2 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.0 | 0.3 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 0.2 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.0 | 0.7 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.4 | GO:2000047 | regulation of cell-cell adhesion mediated by cadherin(GO:2000047) |
| 0.0 | 0.2 | GO:1902292 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.0 | 0.5 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.2 | GO:0006537 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.0 | 0.1 | GO:1903358 | regulation of Golgi organization(GO:1903358) |
| 0.0 | 0.2 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.4 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.2 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.0 | 0.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.2 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.0 | 0.5 | GO:0060034 | notochord cell differentiation(GO:0060034) |
| 0.0 | 0.3 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.4 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.9 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.0 | 0.2 | GO:0006536 | glutamate metabolic process(GO:0006536) |
| 0.0 | 0.1 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.2 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.0 | 0.3 | GO:1901663 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.1 | GO:0032048 | cardiolipin metabolic process(GO:0032048) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.0 | 0.1 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.0 | 0.5 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.1 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.1 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.1 | GO:0051145 | smooth muscle cell differentiation(GO:0051145) negative regulation of muscle cell differentiation(GO:0051148) |
| 0.0 | 0.2 | GO:0045661 | regulation of myoblast differentiation(GO:0045661) |
| 0.0 | 0.2 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.1 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.5 | GO:0009142 | nucleoside triphosphate biosynthetic process(GO:0009142) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.2 | 0.7 | GO:0044279 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.2 | 0.8 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.1 | 0.6 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 0.8 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.8 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 0.5 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.2 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.1 | 0.5 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.5 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:0090443 | FAR/SIN/STRIPAK complex(GO:0090443) |
| 0.0 | 0.5 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.2 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.9 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.2 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.7 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.6 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.2 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0004031 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.1 | 0.8 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.3 | GO:0038046 | enkephalin receptor activity(GO:0038046) |
| 0.1 | 0.3 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.1 | 0.3 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 0.9 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.1 | 0.3 | GO:0048407 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.4 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.1 | 0.3 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 1.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.2 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) protein histidine phosphatase activity(GO:0101006) |
| 0.0 | 0.4 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.3 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.1 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.0 | 0.5 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.2 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.0 | 0.2 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.3 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.2 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.6 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.5 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.1 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.0 | 0.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.2 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.5 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.6 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 0.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.5 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.2 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.0 | 0.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.5 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 1.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.3 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.2 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.0 | 0.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.0 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 0.8 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 0.3 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.7 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.4 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.2 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.5 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.1 | 1.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.4 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.4 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.3 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.7 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.0 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.1 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.4 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |