Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
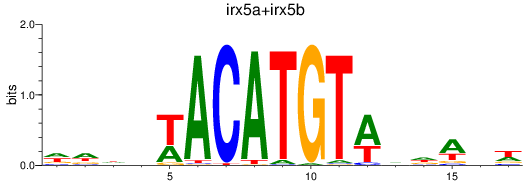
Results for irx5a+irx5b_irx3a+irx3b
Z-value: 2.33
Transcription factors associated with irx5a+irx5b_irx3a+irx3b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
irx5a
|
ENSDARG00000034043 | iroquois homeobox 5a |
|
irx5b
|
ENSDARG00000074070 | iroquois homeobox 5b |
|
irx3b
|
ENSDARG00000031138 | iroquois homeobox 3b |
|
irx3a
|
ENSDARG00000101076 | iroquois homeobox 3a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| irx5a | dr11_v1_chr7_-_35710263_35710263 | -0.94 | 1.3e-04 | Click! |
| irx5b | dr11_v1_chr25_+_36152215_36152215 | -0.92 | 4.2e-04 | Click! |
| irx3a | dr11_v1_chr7_+_36035432_36035432 | -0.89 | 1.4e-03 | Click! |
| irx3b | dr11_v1_chr25_-_36492779_36492779 | -0.77 | 1.6e-02 | Click! |
Activity profile of irx5a+irx5b_irx3a+irx3b motif
Sorted Z-values of irx5a+irx5b_irx3a+irx3b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_-_44574059 | 3.79 |
ENSDART00000123007
|
si:ch73-160p18.3
|
si:ch73-160p18.3 |
| chr15_-_35126332 | 3.51 |
ENSDART00000007636
|
zgc:55413
|
zgc:55413 |
| chr17_+_24809221 | 3.32 |
ENSDART00000082251
ENSDART00000147871 ENSDART00000130871 |
spdya
|
speedy/RINGO cell cycle regulator family member A |
| chr7_+_34587081 | 3.01 |
ENSDART00000173817
|
fhod1
|
formin homology 2 domain containing 1 |
| chr12_+_20693743 | 2.92 |
ENSDART00000153023
ENSDART00000153370 |
st6galnac1.2
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1, tandem duplicate 2 |
| chr2_+_205763 | 2.71 |
ENSDART00000160164
ENSDART00000101071 |
zgc:113293
|
zgc:113293 |
| chr16_-_47381519 | 2.61 |
ENSDART00000032188
ENSDART00000150136 |
si:dkey-256h2.1
|
si:dkey-256h2.1 |
| chr17_-_36860265 | 2.53 |
ENSDART00000182786
ENSDART00000193967 |
senp6b
|
SUMO1/sentrin specific peptidase 6b |
| chr23_+_45845159 | 2.42 |
ENSDART00000023944
|
lmnl3
|
lamin L3 |
| chr17_+_37310663 | 2.37 |
ENSDART00000157122
|
elmsan1b
|
ELM2 and Myb/SANT-like domain containing 1b |
| chr8_-_20914829 | 2.37 |
ENSDART00000025356
|
haus5
|
HAUS augmin-like complex, subunit 5 |
| chr20_-_27190393 | 2.18 |
ENSDART00000149024
|
btbd7
|
BTB (POZ) domain containing 7 |
| chr19_+_32257472 | 2.08 |
ENSDART00000186471
|
atxn1a
|
ataxin 1a |
| chr6_+_33076839 | 2.04 |
ENSDART00000073755
ENSDART00000122242 |
pomgnt1
|
protein O-linked mannose N-acetylglucosaminyltransferase 1 (beta 1,2-) |
| chr9_-_2945008 | 2.04 |
ENSDART00000183452
|
zak
|
sterile alpha motif and leucine zipper containing kinase AZK |
| chr4_-_7869731 | 1.98 |
ENSDART00000067339
|
mcm10
|
minichromosome maintenance 10 replication initiation factor |
| chr7_+_19600262 | 1.96 |
ENSDART00000007310
|
zgc:171731
|
zgc:171731 |
| chr15_+_29472065 | 1.92 |
ENSDART00000154343
|
gdpd5b
|
glycerophosphodiester phosphodiesterase domain containing 5b |
| chr6_+_18441331 | 1.90 |
ENSDART00000171005
|
si:dkey-31g6.4
|
si:dkey-31g6.4 |
| chr5_+_66170479 | 1.90 |
ENSDART00000172117
|
gldc
|
glycine dehydrogenase (decarboxylating) |
| chr10_-_22912255 | 1.89 |
ENSDART00000131992
|
si:ch1073-143l10.2
|
si:ch1073-143l10.2 |
| chr20_+_54299419 | 1.87 |
ENSDART00000056089
ENSDART00000193107 |
si:zfos-1505d6.3
|
si:zfos-1505d6.3 |
| chr21_+_12036238 | 1.84 |
ENSDART00000102463
ENSDART00000155426 |
zgc:162344
|
zgc:162344 |
| chr24_+_12835935 | 1.81 |
ENSDART00000114762
|
nanog
|
nanog homeobox |
| chr25_-_29072162 | 1.80 |
ENSDART00000169269
|
arid3b
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr17_+_14965570 | 1.80 |
ENSDART00000066604
|
gpr137c
|
G protein-coupled receptor 137c |
| chr23_+_45845423 | 1.79 |
ENSDART00000183404
|
lmnl3
|
lamin L3 |
| chr16_+_25116827 | 1.78 |
ENSDART00000163244
|
si:ch211-261d7.6
|
si:ch211-261d7.6 |
| chr3_-_26190804 | 1.77 |
ENSDART00000136001
|
ypel3
|
yippee-like 3 |
| chr16_+_53259521 | 1.68 |
ENSDART00000155461
|
si:ch211-269k10.5
|
si:ch211-269k10.5 |
| chr3_+_43774369 | 1.68 |
ENSDART00000157964
|
zc3h7a
|
zinc finger CCCH-type containing 7A |
| chr21_-_30082414 | 1.67 |
ENSDART00000157307
ENSDART00000155188 |
ccnjl
|
cyclin J-like |
| chr12_+_17154655 | 1.64 |
ENSDART00000028003
|
ankrd22
|
ankyrin repeat domain 22 |
| chr10_+_37400838 | 1.59 |
ENSDART00000136554
|
akap1a
|
A kinase (PRKA) anchor protein 1a |
| chr19_-_2085027 | 1.58 |
ENSDART00000063615
|
snx13
|
sorting nexin 13 |
| chr3_+_33440615 | 1.58 |
ENSDART00000146005
|
gtpbp1
|
GTP binding protein 1 |
| chr15_+_17100412 | 1.58 |
ENSDART00000154418
|
relb
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr8_+_3431671 | 1.57 |
ENSDART00000017850
|
ctu1
|
cytosolic thiouridylase subunit 1 homolog (S. pombe) |
| chr5_-_66429357 | 1.56 |
ENSDART00000160189
|
hip1rb
|
huntingtin interacting protein 1 related b |
| chr18_-_12957451 | 1.55 |
ENSDART00000140403
|
srgap1a
|
SLIT-ROBO Rho GTPase activating protein 1a |
| chr12_+_13091842 | 1.49 |
ENSDART00000185477
ENSDART00000181435 ENSDART00000124799 |
si:ch211-103b1.2
|
si:ch211-103b1.2 |
| chr15_+_28175638 | 1.49 |
ENSDART00000037119
|
slc46a1
|
solute carrier family 46 (folate transporter), member 1 |
| chr7_+_52712807 | 1.48 |
ENSDART00000174095
ENSDART00000174377 ENSDART00000174061 ENSDART00000174094 ENSDART00000110906 ENSDART00000174071 ENSDART00000174238 |
znf280d
|
zinc finger protein 280D |
| chr14_-_26392146 | 1.48 |
ENSDART00000037999
|
b4galt7
|
xylosylprotein beta 1,4-galactosyltransferase, polypeptide 7 (galactosyltransferase I) |
| chr2_+_29995590 | 1.47 |
ENSDART00000151906
|
rbm33b
|
RNA binding motif protein 33b |
| chr4_-_77116266 | 1.45 |
ENSDART00000174249
|
CU467646.2
|
|
| chr20_+_54034512 | 1.44 |
ENSDART00000173226
|
si:dkey-241l7.2
|
si:dkey-241l7.2 |
| chr18_+_25546227 | 1.44 |
ENSDART00000085824
|
pex11a
|
peroxisomal biogenesis factor 11 alpha |
| chr6_+_30113750 | 1.43 |
ENSDART00000022586
|
lrrc40
|
leucine rich repeat containing 40 |
| chr15_-_35112937 | 1.42 |
ENSDART00000154565
ENSDART00000099642 |
zgc:77118
|
zgc:77118 |
| chr7_+_34963748 | 1.42 |
ENSDART00000085087
|
ddx28
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 28 |
| chr22_+_2431585 | 1.42 |
ENSDART00000167758
|
zgc:171435
|
zgc:171435 |
| chr24_-_33308045 | 1.41 |
ENSDART00000149711
|
slc4a2b
|
solute carrier family 4 (anion exchanger), member 2b |
| chr24_+_32525146 | 1.41 |
ENSDART00000132417
ENSDART00000110185 |
yme1l1a
|
YME1-like 1a |
| chr20_+_54018005 | 1.41 |
ENSDART00000123026
ENSDART00000152894 |
si:dkey-241l7.5
|
si:dkey-241l7.5 |
| chr20_+_733510 | 1.41 |
ENSDART00000135066
ENSDART00000015558 ENSDART00000152782 |
myo6a
|
myosin VIa |
| chr6_-_40448416 | 1.40 |
ENSDART00000187019
ENSDART00000179469 |
tatdn2
|
TatD DNase domain containing 2 |
| chr4_-_77125693 | 1.39 |
ENSDART00000174256
|
CU467646.3
|
|
| chr11_-_8126223 | 1.38 |
ENSDART00000091617
ENSDART00000192391 ENSDART00000101561 |
ttll7
|
tubulin tyrosine ligase-like family, member 7 |
| chr20_-_18536474 | 1.36 |
ENSDART00000190903
|
cdc42bpb
|
CDC42 binding protein kinase beta (DMPK-like) |
| chr13_-_86847 | 1.34 |
ENSDART00000158062
|
pole2
|
polymerase (DNA directed), epsilon 2 |
| chr24_-_15263142 | 1.34 |
ENSDART00000183176
ENSDART00000006930 |
rttn
|
rotatin |
| chr19_-_3876877 | 1.34 |
ENSDART00000163711
|
thrap3b
|
thyroid hormone receptor associated protein 3b |
| chr17_+_22311413 | 1.30 |
ENSDART00000151929
|
slc8a1b
|
solute carrier family 8 (sodium/calcium exchanger), member 1b |
| chr11_-_43473824 | 1.29 |
ENSDART00000179561
|
tmem63bb
|
transmembrane protein 63Bb |
| chr9_+_23748133 | 1.29 |
ENSDART00000180758
|
faima
|
Fas apoptotic inhibitory molecule a |
| chr22_-_5724085 | 1.27 |
ENSDART00000110526
ENSDART00000140905 |
pgbd4
|
piggyBac transposable element derived 4 |
| chr3_-_402714 | 1.26 |
ENSDART00000134062
ENSDART00000105659 |
mhc1zja
|
major histocompatibility complex class I ZJA |
| chr7_+_26649319 | 1.26 |
ENSDART00000173823
ENSDART00000101053 |
tp53i11a
|
tumor protein p53 inducible protein 11a |
| chr15_-_31265375 | 1.26 |
ENSDART00000086592
|
vezf1b
|
vascular endothelial zinc finger 1b |
| chr20_+_54024559 | 1.26 |
ENSDART00000130767
|
si:dkey-241l7.4
|
si:dkey-241l7.4 |
| chr4_-_12781182 | 1.25 |
ENSDART00000058020
|
helb
|
helicase (DNA) B |
| chr1_-_34450622 | 1.24 |
ENSDART00000083736
|
lmo7b
|
LIM domain 7b |
| chr20_-_48407944 | 1.24 |
ENSDART00000085624
|
ipo13
|
importin 13 |
| chr13_+_22717366 | 1.24 |
ENSDART00000134122
|
nfkb2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr20_+_54027943 | 1.23 |
ENSDART00000153400
ENSDART00000152961 |
si:dkey-241l7.3
|
si:dkey-241l7.3 |
| chr4_-_5691257 | 1.23 |
ENSDART00000110497
|
tmem63a
|
transmembrane protein 63A |
| chr5_-_48680580 | 1.22 |
ENSDART00000031194
|
lysmd3
|
LysM, putative peptidoglycan-binding, domain containing 3 |
| chr12_+_22674030 | 1.22 |
ENSDART00000153254
ENSDART00000152930 |
cdca9
|
cell division cycle associated 9 |
| chr10_-_28117740 | 1.22 |
ENSDART00000134491
|
med13a
|
mediator complex subunit 13a |
| chr12_+_47448558 | 1.22 |
ENSDART00000185689
ENSDART00000105328 |
fmn2b
|
formin 2b |
| chr20_+_54024724 | 1.22 |
ENSDART00000183562
ENSDART00000152871 |
si:dkey-241l7.4
|
si:dkey-241l7.4 |
| chr4_-_77120928 | 1.21 |
ENSDART00000174154
|
CU467646.1
|
|
| chr8_+_42917515 | 1.21 |
ENSDART00000021715
|
slc23a2
|
solute carrier family 23 (ascorbic acid transporter), member 2 |
| chr18_-_38244871 | 1.19 |
ENSDART00000076399
|
nat10
|
N-acetyltransferase 10 |
| chr16_-_13388821 | 1.19 |
ENSDART00000144062
|
grin2db
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2D, b |
| chr17_-_35076730 | 1.19 |
ENSDART00000146590
|
mboat2a
|
membrane bound O-acyltransferase domain containing 2a |
| chr3_-_43954343 | 1.19 |
ENSDART00000157580
|
ubfd1
|
ubiquitin family domain containing 1 |
| chr18_+_19990412 | 1.17 |
ENSDART00000155054
ENSDART00000090310 |
pias1b
|
protein inhibitor of activated STAT, 1b |
| chr9_+_426392 | 1.17 |
ENSDART00000172515
|
bzw1b
|
basic leucine zipper and W2 domains 1b |
| chr20_+_54034677 | 1.16 |
ENSDART00000173317
ENSDART00000173215 |
si:dkey-241l7.2
|
si:dkey-241l7.2 |
| chr8_+_23034718 | 1.16 |
ENSDART00000184512
|
ythdf1
|
YTH N(6)-methyladenosine RNA binding protein 1 |
| chr20_+_54027778 | 1.16 |
ENSDART00000060458
|
si:dkey-241l7.3
|
si:dkey-241l7.3 |
| chr15_+_19324697 | 1.16 |
ENSDART00000022015
|
vps26b
|
VPS26 retromer complex component B |
| chr9_-_20853439 | 1.15 |
ENSDART00000028247
ENSDART00000133321 |
gdap2
|
ganglioside induced differentiation associated protein 2 |
| chr21_+_13244450 | 1.14 |
ENSDART00000146062
|
specc1lb
|
sperm antigen with calponin homology and coiled-coil domains 1-like b |
| chr10_-_42237304 | 1.14 |
ENSDART00000140341
|
tcf7l1a
|
transcription factor 7 like 1a |
| chr11_-_35970966 | 1.14 |
ENSDART00000188805
|
itpr1b
|
inositol 1,4,5-trisphosphate receptor, type 1b |
| chr1_+_50639416 | 1.14 |
ENSDART00000141977
|
herc3
|
HECT and RLD domain containing E3 ubiquitin protein ligase 3 |
| chr18_-_38245062 | 1.13 |
ENSDART00000189092
|
nat10
|
N-acetyltransferase 10 |
| chr16_+_16266428 | 1.13 |
ENSDART00000188433
|
setd2
|
SET domain containing 2 |
| chr8_+_42941555 | 1.12 |
ENSDART00000183206
|
slc23a2
|
solute carrier family 23 (ascorbic acid transporter), member 2 |
| chr5_-_29152457 | 1.12 |
ENSDART00000078469
|
noxa1
|
NADPH oxidase activator 1 |
| chr11_-_42134968 | 1.11 |
ENSDART00000187115
|
FP325130.1
|
|
| chr21_-_45871866 | 1.11 |
ENSDART00000161716
|
larp1
|
La ribonucleoprotein domain family, member 1 |
| chr20_-_25644131 | 1.10 |
ENSDART00000138997
|
si:dkeyp-117h8.4
|
si:dkeyp-117h8.4 |
| chr1_+_58602506 | 1.10 |
ENSDART00000158425
|
si:ch73-221f6.1
|
si:ch73-221f6.1 |
| chr8_-_53044300 | 1.10 |
ENSDART00000191653
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr17_-_13072334 | 1.10 |
ENSDART00000159598
|
CU469462.1
|
|
| chr13_+_12801092 | 1.09 |
ENSDART00000164040
ENSDART00000134725 |
si:ch211-233a24.2
|
si:ch211-233a24.2 |
| chr21_+_3796196 | 1.09 |
ENSDART00000146754
|
spout1
|
SPOUT domain containing methyltransferase 1 |
| chr25_-_8201983 | 1.08 |
ENSDART00000006579
|
saal1
|
serum amyloid A-like 1 |
| chr22_-_22340688 | 1.08 |
ENSDART00000105597
|
si:ch211-129c21.1
|
si:ch211-129c21.1 |
| chr12_+_34854562 | 1.08 |
ENSDART00000130366
|
si:dkey-21c1.4
|
si:dkey-21c1.4 |
| chr1_+_10127163 | 1.07 |
ENSDART00000110698
|
rbm46
|
RNA binding motif protein 46 |
| chr3_-_40202607 | 1.07 |
ENSDART00000074757
|
znf598
|
zinc finger protein 598 |
| chr8_+_37749263 | 1.06 |
ENSDART00000108556
ENSDART00000147942 |
npm2a
|
nucleophosmin/nucleoplasmin, 2a |
| chr11_-_37880492 | 1.06 |
ENSDART00000102868
|
etnk2
|
ethanolamine kinase 2 |
| chr16_-_7828838 | 1.06 |
ENSDART00000191434
ENSDART00000108653 |
tcaim
|
T cell activation inhibitor, mitochondrial |
| chr7_-_64770456 | 1.06 |
ENSDART00000192618
|
zdhhc21
|
zinc finger, DHHC-type containing 21 |
| chr5_-_58996324 | 1.05 |
ENSDART00000033923
|
mis12
|
MIS12 kinetochore complex component |
| chr6_-_34838397 | 1.05 |
ENSDART00000060169
ENSDART00000169605 |
mier1a
|
mesoderm induction early response 1a, transcriptional regulator |
| chr9_-_41040492 | 1.05 |
ENSDART00000163164
|
adat3
|
adenosine deaminase, tRNA-specific 3 |
| chr3_+_43102010 | 1.04 |
ENSDART00000162096
|
micall2a
|
mical-like 2a |
| chr22_-_23781083 | 1.04 |
ENSDART00000166563
ENSDART00000170458 ENSDART00000166158 ENSDART00000171246 |
cfhl3
|
complement factor H like 3 |
| chr17_-_5352924 | 1.04 |
ENSDART00000167275
|
supt3h
|
SPT3 homolog, SAGA and STAGA complex component |
| chr3_+_27606505 | 1.04 |
ENSDART00000150953
|
usp7
|
ubiquitin specific peptidase 7 (herpes virus-associated) |
| chr16_-_16522013 | 1.03 |
ENSDART00000160602
|
nbeal2
|
neurobeachin-like 2 |
| chr14_-_10617127 | 1.02 |
ENSDART00000154299
|
si:dkey-92i17.2
|
si:dkey-92i17.2 |
| chr1_-_34450784 | 1.02 |
ENSDART00000140515
|
lmo7b
|
LIM domain 7b |
| chr24_-_33366188 | 1.00 |
ENSDART00000074161
|
slc4a2b
|
solute carrier family 4 (anion exchanger), member 2b |
| chr18_+_34181655 | 1.00 |
ENSDART00000130831
ENSDART00000109535 |
gmps
|
guanine monophosphate synthase |
| chr7_-_18877109 | 1.00 |
ENSDART00000113593
|
mllt3
|
MLLT3, super elongation complex subunit |
| chr2_+_27830436 | 1.00 |
ENSDART00000182253
|
FO834800.2
|
|
| chr23_+_34321237 | 1.00 |
ENSDART00000173272
|
plxna1a
|
plexin A1a |
| chr25_+_4855549 | 1.00 |
ENSDART00000163839
|
ap4e1
|
adaptor-related protein complex 4, epsilon 1 subunit |
| chr7_-_6357952 | 0.99 |
ENSDART00000173197
|
zgc:165555
|
zgc:165555 |
| chr25_+_36292465 | 0.98 |
ENSDART00000152649
|
bmb
|
brambleberry |
| chr12_-_33789006 | 0.97 |
ENSDART00000034550
|
llgl2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr6_+_32382743 | 0.97 |
ENSDART00000190009
|
dock7
|
dedicator of cytokinesis 7 |
| chr5_+_7279104 | 0.97 |
ENSDART00000190014
|
si:ch73-72b7.1
|
si:ch73-72b7.1 |
| chr12_+_16087077 | 0.96 |
ENSDART00000141898
|
znf281b
|
zinc finger protein 281b |
| chr17_+_12075805 | 0.96 |
ENSDART00000155329
|
cnsta
|
consortin, connexin sorting protein a |
| chr3_-_45822914 | 0.96 |
ENSDART00000155879
|
si:ch211-66h3.4
|
si:ch211-66h3.4 |
| chr12_-_5448993 | 0.95 |
ENSDART00000181802
|
tbc1d12b
|
TBC1 domain family, member 12b |
| chr5_-_15264007 | 0.95 |
ENSDART00000180641
|
gnb1l
|
guanine nucleotide binding protein (G protein), beta polypeptide 1-like |
| chr9_-_4018452 | 0.94 |
ENSDART00000181059
|
ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr16_+_3067134 | 0.92 |
ENSDART00000012048
|
cyb5r4
|
cytochrome b5 reductase 4 |
| chr13_-_24825691 | 0.92 |
ENSDART00000142745
|
slka
|
STE20-like kinase a |
| chr18_+_14633974 | 0.92 |
ENSDART00000133834
|
vps9d1
|
VPS9 domain containing 1 |
| chr8_-_1219815 | 0.92 |
ENSDART00000016800
ENSDART00000149969 |
znf367
|
zinc finger protein 367 |
| chr1_-_52210950 | 0.91 |
ENSDART00000083946
|
pld6
|
phospholipase D family, member 6 |
| chr2_-_37532772 | 0.90 |
ENSDART00000133951
|
arhgef18a
|
rho/rac guanine nucleotide exchange factor (GEF) 18a |
| chr2_-_57900430 | 0.90 |
ENSDART00000132245
ENSDART00000140060 |
si:dkeyp-68b7.7
|
si:dkeyp-68b7.7 |
| chr8_-_4760723 | 0.90 |
ENSDART00000064201
|
cdc45
|
CDC45 cell division cycle 45 homolog (S. cerevisiae) |
| chr8_-_40442979 | 0.89 |
ENSDART00000157698
ENSDART00000192657 |
tbc1d10aa
|
TBC1 domain family, member 10Aa |
| chr4_+_10604110 | 0.89 |
ENSDART00000122636
ENSDART00000037140 |
ing3
|
inhibitor of growth family, member 3 |
| chr4_+_20486041 | 0.89 |
ENSDART00000017572
|
ints13
|
integrator complex subunit 13 |
| chr11_-_6989598 | 0.89 |
ENSDART00000102493
ENSDART00000173242 ENSDART00000172896 |
zgc:173548
|
zgc:173548 |
| chr25_-_34723937 | 0.89 |
ENSDART00000187286
ENSDART00000157021 |
nup160
|
nucleoporin 160 |
| chr17_+_51746830 | 0.89 |
ENSDART00000184230
|
odc1
|
ornithine decarboxylase 1 |
| chr4_-_6416414 | 0.88 |
ENSDART00000191136
|
MDFIC
|
si:ch73-156e19.1 |
| chr16_-_24642814 | 0.88 |
ENSDART00000153987
ENSDART00000154319 |
fxyd6l
|
FXYD domain containing ion transport regulator 6 like |
| chr7_+_70338270 | 0.87 |
ENSDART00000065234
|
gba3
|
glucosidase, beta, acid 3 (gene/pseudogene) |
| chr21_-_3844322 | 0.87 |
ENSDART00000166652
|
st6galnac4
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 |
| chr9_-_1484202 | 0.87 |
ENSDART00000181215
|
rbm45
|
RNA binding motif protein 45 |
| chr8_-_11832771 | 0.87 |
ENSDART00000193412
ENSDART00000184720 ENSDART00000139947 |
rapgef1a
|
Rap guanine nucleotide exchange factor (GEF) 1a |
| chr7_-_6368406 | 0.86 |
ENSDART00000172787
|
hist1h2a11
|
histone cluster 1 H2A family member 11 |
| chr19_-_27827744 | 0.86 |
ENSDART00000181620
|
papd7
|
PAP associated domain containing 7 |
| chr6_+_49901465 | 0.86 |
ENSDART00000023515
|
chmp4ba
|
charged multivesicular body protein 4Ba |
| chr17_+_28675120 | 0.85 |
ENSDART00000159067
|
hectd1
|
HECT domain containing 1 |
| chr1_+_51191049 | 0.85 |
ENSDART00000132244
ENSDART00000014970 ENSDART00000132141 |
btbd3a
|
BTB (POZ) domain containing 3a |
| chr14_-_8940499 | 0.85 |
ENSDART00000129030
|
zgc:153681
|
zgc:153681 |
| chr16_+_16265850 | 0.85 |
ENSDART00000181265
|
setd2
|
SET domain containing 2 |
| chr19_+_26340736 | 0.84 |
ENSDART00000013497
|
mylipa
|
myosin regulatory light chain interacting protein a |
| chr23_+_42131509 | 0.83 |
ENSDART00000184544
|
SHISAL2A
|
shisa like 2A |
| chr25_-_36263115 | 0.83 |
ENSDART00000143046
ENSDART00000139002 |
dus2
|
dihydrouridine synthase 2 |
| chr4_-_20313810 | 0.83 |
ENSDART00000136350
|
dcp1b
|
decapping mRNA 1B |
| chr24_-_26981848 | 0.83 |
ENSDART00000183198
|
stag1b
|
stromal antigen 1b |
| chr19_-_7043355 | 0.83 |
ENSDART00000104845
|
tapbp.1
|
TAP binding protein (tapasin), tandem duplicate 1 |
| chr25_-_17590971 | 0.82 |
ENSDART00000189942
|
mmp15a
|
matrix metallopeptidase 15a |
| chr13_+_24671481 | 0.82 |
ENSDART00000001678
|
adam8a
|
ADAM metallopeptidase domain 8a |
| chr25_+_6266009 | 0.82 |
ENSDART00000148995
|
slc25a44a
|
solute carrier family 25, member 44 a |
| chr23_-_3721444 | 0.81 |
ENSDART00000141682
|
nudt3a
|
nudix (nucleoside diphosphate linked moiety X)-type motif 3a |
| chr9_+_24088062 | 0.81 |
ENSDART00000126198
|
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr6_-_28961660 | 0.81 |
ENSDART00000147285
|
tomm70a
|
translocase of outer mitochondrial membrane 70 homolog A (S. cerevisiae) |
| chr3_+_18050667 | 0.81 |
ENSDART00000035531
|
mettl26
|
methyltransferase like 26 |
| chr13_+_6092135 | 0.80 |
ENSDART00000162738
|
fam120b
|
family with sequence similarity 120B |
| chr16_+_29516098 | 0.80 |
ENSDART00000174895
|
ctss2.2
|
cathepsin S, ortholog 2, tandem duplicate 2 |
| chr23_-_33680265 | 0.80 |
ENSDART00000138416
|
tfcp2
|
transcription factor CP2 |
| chr23_+_29885019 | 0.79 |
ENSDART00000167059
|
aurkaip1
|
aurora kinase A interacting protein 1 |
| chr2_-_29923403 | 0.79 |
ENSDART00000144672
|
paxip1
|
PAX interacting (with transcription-activation domain) protein 1 |
| chr13_-_25719628 | 0.79 |
ENSDART00000135383
|
si:dkey-192p21.6
|
si:dkey-192p21.6 |
| chr18_-_44847855 | 0.79 |
ENSDART00000086823
|
srpr
|
signal recognition particle receptor (docking protein) |
| chr2_-_43850897 | 0.79 |
ENSDART00000005449
|
zeb1a
|
zinc finger E-box binding homeobox 1a |
| chr23_-_19977214 | 0.79 |
ENSDART00000054659
|
ccnq
|
cyclin Q |
| chr14_+_17397534 | 0.79 |
ENSDART00000180162
ENSDART00000129838 |
rnf212
|
ring finger protein 212 |
| chr4_-_9191220 | 0.79 |
ENSDART00000156919
|
hcfc2
|
host cell factor C2 |
| chr1_+_46404968 | 0.79 |
ENSDART00000042064
|
tubgcp3
|
tubulin, gamma complex associated protein 3 |
| chr13_-_24745288 | 0.78 |
ENSDART00000031564
|
sfr1
|
SWI5-dependent homologous recombination repair protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of irx5a+irx5b_irx3a+irx3b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0015882 | L-ascorbic acid transport(GO:0015882) L-ascorbic acid metabolic process(GO:0019852) |
| 0.7 | 2.0 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.6 | 2.5 | GO:0090230 | regulation of centromere complex assembly(GO:0090230) regulation of kinetochore assembly(GO:0090234) |
| 0.5 | 1.6 | GO:0010657 | negative regulation of muscle cell apoptotic process(GO:0010656) muscle cell apoptotic process(GO:0010657) striated muscle cell apoptotic process(GO:0010658) regulation of muscle cell apoptotic process(GO:0010660) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 0.5 | 1.9 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.4 | 1.1 | GO:0097095 | frontonasal suture morphogenesis(GO:0097095) |
| 0.4 | 1.9 | GO:0039689 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.4 | 1.1 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.3 | 2.0 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 0.3 | 1.0 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.3 | 0.9 | GO:1902001 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.3 | 1.3 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.3 | 2.0 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.3 | 1.7 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.3 | 1.1 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.3 | 1.4 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.3 | 0.8 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.3 | 1.1 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.3 | 1.1 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.3 | 1.6 | GO:0032447 | tRNA wobble position uridine thiolation(GO:0002143) protein urmylation(GO:0032447) |
| 0.3 | 1.0 | GO:0036344 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.2 | 0.7 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.2 | 0.7 | GO:0010526 | regulation of transposition, RNA-mediated(GO:0010525) negative regulation of transposition, RNA-mediated(GO:0010526) transposition, RNA-mediated(GO:0032197) positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 0.2 | 1.4 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.2 | 0.7 | GO:0010452 | histone H3-K36 methylation(GO:0010452) |
| 0.2 | 0.7 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.2 | 1.1 | GO:0010692 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.2 | 1.9 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.2 | 0.6 | GO:1902767 | farnesyl diphosphate biosynthetic process, mevalonate pathway(GO:0010142) isoprenoid biosynthetic process via mevalonate(GO:1902767) |
| 0.2 | 1.0 | GO:0000741 | karyogamy(GO:0000741) pronuclear fusion(GO:0007344) |
| 0.2 | 0.9 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.2 | 2.0 | GO:1902041 | regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902041) negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.2 | 1.5 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.2 | 0.8 | GO:1901908 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.2 | 0.5 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.2 | 0.3 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.2 | 0.5 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.1 | 1.8 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.1 | 0.4 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.1 | 0.7 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.1 | 1.7 | GO:0035196 | production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.1 | 1.1 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.1 | 0.6 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.1 | 0.8 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 2.3 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.1 | 2.1 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 3.0 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.5 | GO:0030890 | B cell homeostasis(GO:0001782) positive regulation of B cell proliferation(GO:0030890) |
| 0.1 | 0.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 0.5 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.1 | 1.8 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 | 0.7 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 1.7 | GO:0000479 | endonucleolytic cleavage involved in rRNA processing(GO:0000478) endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.1 | 0.9 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.1 | 2.0 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.6 | GO:0070587 | regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 0.1 | 1.0 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 1.4 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 0.5 | GO:0045843 | negative regulation of striated muscle tissue development(GO:0045843) negative regulation of muscle tissue development(GO:1901862) |
| 0.1 | 0.7 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 0.6 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) |
| 0.1 | 0.9 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.1 | 0.8 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.1 | 0.5 | GO:2000781 | positive regulation of double-strand break repair(GO:2000781) |
| 0.1 | 2.5 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 1.1 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.1 | 0.5 | GO:0071480 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.1 | 0.9 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.1 | 3.2 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.1 | 1.2 | GO:0061157 | RNA destabilization(GO:0050779) mRNA destabilization(GO:0061157) |
| 0.1 | 0.1 | GO:0006168 | adenine salvage(GO:0006168) |
| 0.1 | 0.8 | GO:0055015 | ventricular cardiac muscle cell development(GO:0055015) |
| 0.1 | 0.5 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) mitochondrial DNA replication(GO:0006264) mitochondrial DNA metabolic process(GO:0032042) |
| 0.1 | 1.5 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.1 | 0.9 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.1 | 0.3 | GO:0071586 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.1 | 0.5 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.1 | 1.5 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 0.5 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.1 | 0.4 | GO:1903019 | negative regulation of glycoprotein biosynthetic process(GO:0010561) negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) negative regulation of glycoprotein metabolic process(GO:1903019) |
| 0.1 | 0.4 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.1 | 0.4 | GO:0032782 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.1 | 0.3 | GO:0030238 | female sex determination(GO:0030237) male sex determination(GO:0030238) |
| 0.1 | 0.5 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.1 | 0.7 | GO:0006478 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.1 | 1.0 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 0.7 | GO:0006203 | dGTP catabolic process(GO:0006203) purine nucleoside triphosphate catabolic process(GO:0009146) purine deoxyribonucleoside triphosphate catabolic process(GO:0009217) dGTP metabolic process(GO:0046070) |
| 0.1 | 0.6 | GO:0043201 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.1 | 1.1 | GO:0034244 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.1 | 0.3 | GO:0042256 | mature ribosome assembly(GO:0042256) assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 | 0.4 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.1 | 2.4 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.1 | 0.4 | GO:0014005 | microglia development(GO:0014005) |
| 0.1 | 0.4 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.1 | 1.5 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.1 | 0.5 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.1 | 1.3 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.1 | 0.3 | GO:0032207 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) |
| 0.1 | 1.1 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.1 | 0.8 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 | 0.5 | GO:0003188 | heart valve formation(GO:0003188) atrioventricular valve formation(GO:0003190) |
| 0.1 | 1.0 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.1 | 0.2 | GO:0043455 | regulation of secondary metabolic process(GO:0043455) |
| 0.1 | 0.4 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 | 0.3 | GO:0070199 | establishment of mitotic sister chromatid cohesion(GO:0034087) establishment of protein localization to chromosome(GO:0070199) establishment of protein localization to chromatin(GO:0071169) transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.1 | 0.3 | GO:0070291 | N-acylethanolamine metabolic process(GO:0070291) |
| 0.1 | 0.6 | GO:0044090 | positive regulation of vacuole organization(GO:0044090) |
| 0.1 | 1.0 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.1 | 0.3 | GO:2000815 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.1 | 0.3 | GO:0090076 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) negative regulation of striated muscle contraction(GO:0045988) relaxation of skeletal muscle(GO:0090076) |
| 0.1 | 1.4 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 0.9 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.1 | 0.2 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 0.8 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.1 | 0.6 | GO:0098789 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 0.3 | GO:1902165 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902165) |
| 0.1 | 0.4 | GO:0006596 | polyamine biosynthetic process(GO:0006596) |
| 0.1 | 0.7 | GO:0046037 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.1 | 0.7 | GO:0006032 | chitin catabolic process(GO:0006032) |
| 0.1 | 0.5 | GO:0031106 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.1 | 0.6 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.1 | 0.4 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) |
| 0.1 | 1.4 | GO:0048844 | artery morphogenesis(GO:0048844) |
| 0.1 | 1.2 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.1 | 0.6 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.1 | 0.7 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 0.3 | GO:0097037 | heme export(GO:0097037) |
| 0.1 | 0.9 | GO:0071156 | regulation of cell cycle arrest(GO:0071156) |
| 0.1 | 1.3 | GO:1901888 | regulation of cell junction assembly(GO:1901888) |
| 0.1 | 2.1 | GO:0061138 | morphogenesis of a branching epithelium(GO:0061138) |
| 0.1 | 0.3 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 0.7 | GO:1900052 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.1 | 0.7 | GO:0033198 | response to ATP(GO:0033198) |
| 0.1 | 1.7 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.1 | 0.6 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.1 | 0.4 | GO:0070285 | pigment cell development(GO:0070285) |
| 0.1 | 1.4 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.1 | 1.4 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.1 | 1.7 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 0.2 | GO:1904478 | regulation of intestinal absorption(GO:1904478) regulation of intestinal lipid absorption(GO:1904729) |
| 0.1 | 0.4 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.1 | 3.3 | GO:0007098 | centrosome cycle(GO:0007098) centrosome organization(GO:0051297) |
| 0.1 | 0.6 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.1 | 0.5 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.0 | 0.7 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 2.6 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.2 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.6 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.0 | 0.2 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.0 | 0.6 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.2 | GO:0097510 | base-excision repair, AP site formation via deaminated base removal(GO:0097510) |
| 0.0 | 0.9 | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.5 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 1.5 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.2 | GO:0098529 | neuromuscular junction development, skeletal muscle fiber(GO:0098529) |
| 0.0 | 1.0 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.4 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.1 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.2 | GO:0001836 | release of cytochrome c from mitochondria(GO:0001836) |
| 0.0 | 0.6 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.1 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.0 | 0.4 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.5 | GO:0036353 | histone H2A monoubiquitination(GO:0035518) histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.3 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 0.2 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.0 | 0.4 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 1.1 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.7 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.1 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.8 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.0 | 0.4 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.1 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 0.0 | 0.0 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.0 | 0.3 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.3 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.2 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.0 | 0.6 | GO:0035308 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.0 | 0.4 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.2 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.4 | GO:2001057 | reactive nitrogen species metabolic process(GO:2001057) |
| 0.0 | 0.1 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.4 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 0.2 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.0 | 0.5 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0097053 | L-kynurenine metabolic process(GO:0097052) L-kynurenine catabolic process(GO:0097053) |
| 0.0 | 0.8 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.1 | GO:0090387 | phagosome maturation involved in apoptotic cell clearance(GO:0090386) phagolysosome assembly involved in apoptotic cell clearance(GO:0090387) |
| 0.0 | 0.2 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 1.6 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.3 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.0 | 0.6 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 1.8 | GO:0030166 | proteoglycan biosynthetic process(GO:0030166) |
| 0.0 | 0.4 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.2 | GO:0019563 | glycerol catabolic process(GO:0019563) |
| 0.0 | 0.2 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.1 | GO:1990173 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) protein localization to nucleoplasm(GO:1990173) |
| 0.0 | 0.5 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 1.2 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.7 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.2 | GO:0034080 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.9 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 1.6 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 1.0 | GO:0031647 | regulation of protein stability(GO:0031647) |
| 0.0 | 0.6 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.2 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.5 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 0.2 | GO:0035552 | oxidative single-stranded DNA demethylation(GO:0035552) |
| 0.0 | 0.3 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.8 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.6 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.0 | 2.1 | GO:0000724 | double-strand break repair via homologous recombination(GO:0000724) |
| 0.0 | 0.8 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 2.0 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 1.9 | GO:0034504 | protein localization to nucleus(GO:0034504) |
| 0.0 | 1.4 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.5 | GO:0045841 | mitotic spindle assembly checkpoint(GO:0007094) spindle checkpoint(GO:0031577) negative regulation of mitotic metaphase/anaphase transition(GO:0045841) spindle assembly checkpoint(GO:0071173) mitotic spindle checkpoint(GO:0071174) |
| 0.0 | 0.3 | GO:0051121 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.5 | GO:0007257 | activation of JUN kinase activity(GO:0007257) regulation of JUN kinase activity(GO:0043506) positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 0.8 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 0.9 | GO:0042752 | regulation of circadian rhythm(GO:0042752) |
| 0.0 | 0.2 | GO:0070836 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.5 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.2 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.3 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.0 | 0.5 | GO:0006282 | regulation of DNA repair(GO:0006282) |
| 0.0 | 0.7 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 0.6 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.1 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.3 | GO:0045910 | negative regulation of DNA recombination(GO:0045910) |
| 0.0 | 0.3 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.2 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 1.1 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.4 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 2.2 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.3 | GO:0045920 | negative regulation of exocytosis(GO:0045920) |
| 0.0 | 0.1 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.0 | 0.9 | GO:0000070 | mitotic sister chromatid segregation(GO:0000070) |
| 0.0 | 0.9 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 1.6 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.4 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.0 | 0.4 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.1 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) |
| 0.0 | 0.0 | GO:1903430 | regulation of oocyte maturation(GO:1900193) negative regulation of cell maturation(GO:1903430) |
| 0.0 | 0.7 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.2 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.2 | GO:0045022 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.0 | 1.2 | GO:0032147 | activation of protein kinase activity(GO:0032147) |
| 0.0 | 0.8 | GO:0045010 | actin nucleation(GO:0045010) |
| 0.0 | 0.2 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 1.7 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.6 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 1.1 | GO:0031497 | chromatin assembly(GO:0031497) |
| 0.0 | 0.1 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.5 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.0 | 0.2 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.0 | 0.2 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.0 | 1.1 | GO:0007179 | transforming growth factor beta receptor signaling pathway(GO:0007179) response to transforming growth factor beta(GO:0071559) cellular response to transforming growth factor beta stimulus(GO:0071560) |
| 0.0 | 0.7 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 0.2 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.1 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.8 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.1 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.1 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 1.5 | GO:0006281 | DNA repair(GO:0006281) |
| 0.0 | 0.2 | GO:0060323 | head morphogenesis(GO:0060323) |
| 0.0 | 0.6 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.2 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.9 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.6 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.2 | GO:0021986 | habenula development(GO:0021986) |
| 0.0 | 0.2 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.5 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.3 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.4 | GO:0021549 | cerebellum development(GO:0021549) |
| 0.0 | 0.2 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.0 | 0.9 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.0 | GO:0046931 | pore complex assembly(GO:0046931) |
| 0.0 | 0.4 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.4 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 0.4 | GO:0006626 | protein targeting to mitochondrion(GO:0006626) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.4 | 1.3 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.4 | 2.0 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.4 | 0.4 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.3 | 1.3 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.3 | 0.8 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.3 | 1.1 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.3 | 2.4 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.2 | 1.7 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.2 | 0.7 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.2 | 1.2 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.2 | 1.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.2 | 0.7 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.2 | 1.4 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.2 | 1.0 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.2 | 0.3 | GO:0000941 | condensed nuclear chromosome inner kinetochore(GO:0000941) |
| 0.1 | 1.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 0.6 | GO:0005880 | nuclear microtubule(GO:0005880) |
| 0.1 | 0.6 | GO:1904423 | dehydrodolichyl diphosphate synthase complex(GO:1904423) |
| 0.1 | 1.2 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 0.9 | GO:0031261 | DNA replication preinitiation complex(GO:0031261) |
| 0.1 | 0.8 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 1.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 1.5 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.6 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 0.8 | GO:0033061 | DNA recombinase mediator complex(GO:0033061) |
| 0.1 | 0.4 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 0.9 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.1 | 0.6 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.1 | 0.7 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.1 | 0.4 | GO:0055087 | Ski complex(GO:0055087) |
| 0.1 | 1.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 0.4 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 0.5 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.1 | 0.6 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 0.4 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 0.3 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 0.4 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 1.3 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.1 | 0.5 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 0.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.9 | GO:0044545 | NSL complex(GO:0044545) |
| 0.1 | 1.1 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 1.9 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 0.7 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 1.6 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.1 | 0.5 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.1 | 1.0 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.1 | 0.3 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.1 | 0.2 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 1.1 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 1.3 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.8 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.8 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.4 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 2.4 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.2 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 2.2 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.9 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.9 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.4 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.5 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.6 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 1.0 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 11.2 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.7 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 1.0 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 1.4 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 1.9 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.5 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.4 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.7 | GO:0030118 | clathrin coat(GO:0030118) clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.5 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.2 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.2 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.2 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.0 | 0.2 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 1.0 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 1.3 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.7 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.7 | GO:0071010 | U2-type prespliceosome(GO:0071004) prespliceosome(GO:0071010) |
| 0.0 | 1.9 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.3 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.5 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 3.8 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 0.7 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.2 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 1.0 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 1.8 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.6 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 0.6 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 2.7 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.6 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.5 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.5 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.9 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 1.6 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 2.2 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.3 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 1.3 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 2.2 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.3 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.4 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 1.0 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.7 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.3 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.3 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.5 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 2.2 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 0.1 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.4 | GO:0055029 | nuclear DNA-directed RNA polymerase complex(GO:0055029) |
| 0.0 | 0.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.2 | GO:0098844 | dendritic spine head(GO:0044327) postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.1 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.0 | 0.0 | GO:0044215 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 1.0 | GO:0005681 | spliceosomal complex(GO:0005681) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.6 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.3 | 1.0 | GO:0052717 | tRNA-specific adenosine-34 deaminase activity(GO:0052717) |
| 0.3 | 3.5 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.3 | 2.5 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.3 | 1.5 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.2 | 1.0 | GO:0071253 | connexin binding(GO:0071253) |
| 0.2 | 0.7 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.2 | 1.7 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.2 | 0.6 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.2 | 1.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.2 | 0.6 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.2 | 3.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.2 | 0.7 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
| 0.2 | 0.9 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.2 | 1.9 | GO:0016594 | glycine binding(GO:0016594) |
| 0.2 | 2.7 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.2 | 1.3 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.2 | 0.8 | GO:0008486 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.2 | 0.6 | GO:0031151 | histone methyltransferase activity (H3-K79 specific)(GO:0031151) |
| 0.2 | 1.4 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.2 | 0.6 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.1 | 0.7 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.1 | 0.6 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.1 | 1.6 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.8 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 1.4 | GO:0043142 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.1 | 2.4 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.1 | 0.7 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 0.6 | GO:0008311 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.1 | 1.9 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 0.7 | GO:0001047 | core promoter sequence-specific DNA binding(GO:0001046) core promoter binding(GO:0001047) |
| 0.1 | 0.7 | GO:0016793 | dGTPase activity(GO:0008832) triphosphoric monoester hydrolase activity(GO:0016793) |
| 0.1 | 2.4 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.1 | 1.3 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 1.4 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 3.3 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.1 | 0.6 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 0.3 | GO:0050135 | NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.1 | 0.5 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.1 | 0.9 | GO:0004586 | ornithine decarboxylase activity(GO:0004586) |
| 0.1 | 2.0 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 1.4 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 1.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 2.1 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 0.6 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.3 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.1 | 4.3 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.1 | 0.7 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.1 | 0.4 | GO:0015432 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.1 | 1.6 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 0.7 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.1 | 0.9 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 0.6 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 0.2 | GO:0030623 | U5 snRNA binding(GO:0030623) |
| 0.1 | 0.9 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 1.4 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 0.5 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.1 | 0.7 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 0.2 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.1 | 0.5 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 1.0 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.1 | 0.3 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.1 | 0.5 | GO:0000992 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 3 promoter sequence-specific DNA binding(GO:0001006) RNA polymerase III regulatory region DNA binding(GO:0001016) RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.1 | 0.3 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.1 | 0.7 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.1 | 0.9 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.1 | 0.2 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.1 | 0.6 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.6 | GO:0015924 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.1 | 1.2 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 4.7 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.1 | 0.7 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 1.0 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 2.6 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.1 | 0.6 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) tRNA-specific adenosine deaminase activity(GO:0008251) |
| 0.1 | 0.7 | GO:0005231 | extracellular ATP-gated cation channel activity(GO:0004931) excitatory extracellular ligand-gated ion channel activity(GO:0005231) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 0.4 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 0.2 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.1 | 1.0 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 1.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 0.7 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 0.3 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.1 | 1.8 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 0.2 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.1 | 0.4 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.0 | 0.5 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.8 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 0.2 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.0 | 0.2 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.0 | 0.7 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 1.5 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.3 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.6 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.5 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.9 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.7 | GO:0061608 | nucleocytoplasmic transporter activity(GO:0005487) nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.3 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.5 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 1.1 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.3 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.0 | 0.3 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.8 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.5 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.3 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.2 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.3 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.0 | 0.8 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.3 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.3 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.9 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.4 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 1.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0003999 | adenine phosphoribosyltransferase activity(GO:0003999) |
| 0.0 | 0.4 | GO:0043028 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 0.2 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.5 | GO:0048018 | receptor activator activity(GO:0030546) receptor agonist activity(GO:0048018) |
| 0.0 | 0.2 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.0 | 0.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.3 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.2 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.8 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.4 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.5 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.1 | GO:0016453 | C-acetyltransferase activity(GO:0016453) |
| 0.0 | 0.3 | GO:0016840 | carbon-nitrogen lyase activity(GO:0016840) |
| 0.0 | 0.5 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.3 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.5 | GO:0008173 | RNA methyltransferase activity(GO:0008173) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.2 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 6.4 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.3 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.5 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.4 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.6 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 0.9 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.4 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 1.1 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.3 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.4 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.8 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.2 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.4 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.5 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.2 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.3 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 3.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.3 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.0 | 1.9 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 1.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.4 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.7 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 3.4 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 0.9 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 2.2 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.1 | GO:0019865 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.0 | 0.8 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 1.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 1.0 | GO:0019209 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.0 | 5.8 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.7 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 3.4 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 0.1 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.1 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.0 | 0.3 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.3 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.0 | 0.9 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 0.5 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.5 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.4 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 1.1 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.3 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.6 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.4 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.4 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 6.9 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.3 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.1 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 2.5 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.5 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.5 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.0 | 0.5 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.1 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.4 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.0 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.1 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.0 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 1.6 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 2.5 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 1.8 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 2.5 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 1.7 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 0.4 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 1.6 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 1.1 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.6 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 1.0 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.8 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.9 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.5 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.1 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.7 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.7 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.7 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.4 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.4 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.4 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.4 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.6 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.7 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.4 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.5 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.3 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.4 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.9 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.2 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.2 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.8 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.4 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.4 | PID PLK1 PATHWAY | PLK1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 1.6 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 2.1 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.1 | 3.9 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.1 | 1.0 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 0.9 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 0.5 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 1.0 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.1 | 1.7 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.1 | 1.5 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 1.1 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 0.7 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 0.5 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 0.6 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 0.8 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.1 | 0.8 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 1.2 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 1.4 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.8 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.8 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.7 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.4 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.2 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.0 | 2.3 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.0 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.4 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.8 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.3 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 1.9 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 1.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.4 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 0.7 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.1 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.6 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.2 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.1 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.8 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.4 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 1.3 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.3 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.1 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.8 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |