Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
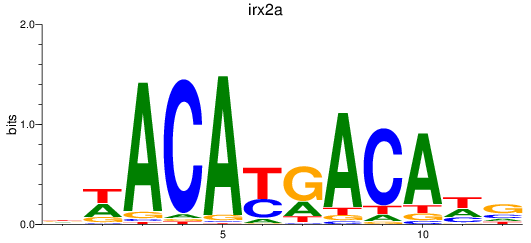
Results for irx2a
Z-value: 0.43
Transcription factors associated with irx2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
irx2a
|
ENSDARG00000001785 | iroquois homeobox 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| irx2a | dr11_v1_chr16_-_560574_560574 | 0.55 | 1.3e-01 | Click! |
Activity profile of irx2a motif
Sorted Z-values of irx2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_+_32499916 | 0.81 |
ENSDART00000134811
|
si:dkey-261h17.1
|
si:dkey-261h17.1 |
| chr7_-_2039060 | 0.72 |
ENSDART00000173879
|
si:cabz01007794.1
|
si:cabz01007794.1 |
| chr11_+_3254252 | 0.63 |
ENSDART00000123568
|
pmela
|
premelanosome protein a |
| chr3_-_61203203 | 0.61 |
ENSDART00000171787
|
pvalb1
|
parvalbumin 1 |
| chr9_+_31282161 | 0.61 |
ENSDART00000010774
|
zic2a
|
zic family member 2 (odd-paired homolog, Drosophila), a |
| chr12_+_36891483 | 0.61 |
ENSDART00000048927
|
cox10
|
COX10 heme A:farnesyltransferase cytochrome c oxidase assembly factor |
| chr1_-_44940830 | 0.60 |
ENSDART00000097500
ENSDART00000134464 ENSDART00000137216 |
tmem176
|
transmembrane protein 176 |
| chr20_+_2281933 | 0.59 |
ENSDART00000137579
|
si:ch73-18b11.2
|
si:ch73-18b11.2 |
| chr19_-_41069573 | 0.54 |
ENSDART00000111982
ENSDART00000193142 |
sgce
|
sarcoglycan, epsilon |
| chr6_-_18698006 | 0.53 |
ENSDART00000170128
|
rhbdl3
|
rhomboid, veinlet-like 3 (Drosophila) |
| chr7_-_27033080 | 0.52 |
ENSDART00000173516
|
nucb2a
|
nucleobindin 2a |
| chr19_+_18739085 | 0.52 |
ENSDART00000188868
|
spaca4l
|
sperm acrosome associated 4 like |
| chr14_-_48103207 | 0.50 |
ENSDART00000056712
|
etfdh
|
electron-transferring-flavoprotein dehydrogenase |
| chr24_-_12938922 | 0.49 |
ENSDART00000024084
|
pck2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial) |
| chr19_+_8973042 | 0.46 |
ENSDART00000039597
|
crabp2b
|
cellular retinoic acid binding protein 2, b |
| chr7_-_73725701 | 0.44 |
ENSDART00000158525
|
casq1b
|
calsequestrin 1b |
| chr21_+_723998 | 0.44 |
ENSDART00000160956
|
oaz1b
|
ornithine decarboxylase antizyme 1b |
| chr8_+_44926946 | 0.43 |
ENSDART00000098567
|
zgc:154046
|
zgc:154046 |
| chr23_+_36653376 | 0.42 |
ENSDART00000053189
|
gpr182
|
G protein-coupled receptor 182 |
| chr24_-_26304386 | 0.42 |
ENSDART00000175416
|
otos
|
otospiralin |
| chr19_+_30388186 | 0.41 |
ENSDART00000103474
|
tspan13b
|
tetraspanin 13b |
| chr19_+_30387999 | 0.41 |
ENSDART00000145396
|
tspan13b
|
tetraspanin 13b |
| chr23_+_42819221 | 0.40 |
ENSDART00000180495
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr24_-_37955993 | 0.40 |
ENSDART00000041805
|
metrn
|
meteorin, glial cell differentiation regulator |
| chr2_+_22795494 | 0.40 |
ENSDART00000042255
|
rab6bb
|
RAB6B, member RAS oncogene family b |
| chr8_+_23213320 | 0.39 |
ENSDART00000032996
ENSDART00000137536 |
ppdpfa
|
pancreatic progenitor cell differentiation and proliferation factor a |
| chr1_+_44941031 | 0.37 |
ENSDART00000141145
|
si:dkey-9i23.16
|
si:dkey-9i23.16 |
| chr8_+_26565512 | 0.36 |
ENSDART00000140980
|
sema3bl
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3bl |
| chr14_-_25985698 | 0.35 |
ENSDART00000172909
ENSDART00000123053 |
atox1
|
antioxidant 1 copper chaperone |
| chr4_-_12725513 | 0.35 |
ENSDART00000132286
|
mgst1.2
|
microsomal glutathione S-transferase 1.2 |
| chr6_-_18698456 | 0.35 |
ENSDART00000166587
|
rhbdl3
|
rhomboid, veinlet-like 3 (Drosophila) |
| chr22_-_21314821 | 0.34 |
ENSDART00000105546
ENSDART00000135388 |
cks2
|
CDC28 protein kinase regulatory subunit 2 |
| chr3_+_32689707 | 0.34 |
ENSDART00000029262
|
si:dkey-16l2.17
|
si:dkey-16l2.17 |
| chr8_-_25011157 | 0.33 |
ENSDART00000078795
|
ahcyl1
|
adenosylhomocysteinase-like 1 |
| chr24_-_7632187 | 0.32 |
ENSDART00000041714
|
atp6v0a1b
|
ATPase H+ transporting V0 subunit a1b |
| chr7_-_23873173 | 0.32 |
ENSDART00000101408
|
zgc:162171
|
zgc:162171 |
| chr9_-_56231387 | 0.32 |
ENSDART00000149851
|
rpl31
|
ribosomal protein L31 |
| chr11_+_44579865 | 0.31 |
ENSDART00000173425
|
nid1b
|
nidogen 1b |
| chr6_-_7720332 | 0.31 |
ENSDART00000135945
|
rpsa
|
ribosomal protein SA |
| chr6_-_19340889 | 0.31 |
ENSDART00000181407
|
mif4gda
|
MIF4G domain containing a |
| chr6_-_19341184 | 0.31 |
ENSDART00000168236
ENSDART00000167674 |
mif4gda
|
MIF4G domain containing a |
| chr21_-_37733287 | 0.30 |
ENSDART00000157826
|
mpp1
|
membrane protein, palmitoylated 1 |
| chr2_+_47581997 | 0.29 |
ENSDART00000112579
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr17_-_35881841 | 0.29 |
ENSDART00000110040
|
sox11a
|
SRY (sex determining region Y)-box 11a |
| chr2_-_10014821 | 0.29 |
ENSDART00000185525
|
CR956623.1
|
|
| chr5_+_37785152 | 0.29 |
ENSDART00000053511
ENSDART00000189812 |
myo1ca
|
myosin Ic, paralog a |
| chr23_-_39849155 | 0.29 |
ENSDART00000115330
|
ppp1r14c
|
protein phosphatase 1, regulatory (inhibitor) subunit 14C |
| chr6_-_18698669 | 0.28 |
ENSDART00000168554
ENSDART00000175205 |
rhbdl3
|
rhomboid, veinlet-like 3 (Drosophila) |
| chr10_-_17083180 | 0.28 |
ENSDART00000170083
|
fam166b
|
family with sequence similarity 166, member B |
| chr14_+_32022272 | 0.28 |
ENSDART00000105760
|
zic6
|
zic family member 6 |
| chr17_+_32571584 | 0.28 |
ENSDART00000087565
|
eva1a
|
eva-1 homolog A (C. elegans) |
| chr11_-_21585176 | 0.28 |
ENSDART00000045391
|
srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr8_-_27858458 | 0.27 |
ENSDART00000132632
ENSDART00000136562 |
cttnbp2nlb
|
CTTNBP2 N-terminal like b |
| chr13_+_18533005 | 0.27 |
ENSDART00000136024
|
ftr14l
|
finTRIM family, member 14-like |
| chr20_+_26349002 | 0.27 |
ENSDART00000152842
|
syne1a
|
spectrin repeat containing, nuclear envelope 1a |
| chr21_-_9991502 | 0.27 |
ENSDART00000169320
|
esm1
|
endothelial cell-specific molecule 1 |
| chr8_-_30791266 | 0.27 |
ENSDART00000062220
|
gstt1a
|
glutathione S-transferase theta 1a |
| chr23_+_23658474 | 0.27 |
ENSDART00000162838
|
agrn
|
agrin |
| chr21_+_5915041 | 0.27 |
ENSDART00000151370
|
prodha
|
proline dehydrogenase (oxidase) 1a |
| chr3_+_62161184 | 0.27 |
ENSDART00000090370
ENSDART00000192665 |
noxo1a
|
NADPH oxidase organizer 1a |
| chr23_-_28239750 | 0.26 |
ENSDART00000003548
|
znf385a
|
zinc finger protein 385A |
| chr21_-_8513192 | 0.26 |
ENSDART00000084378
|
crb2a
|
crumbs family member 2a |
| chr19_+_10832092 | 0.26 |
ENSDART00000191851
|
tomm40l
|
translocase of outer mitochondrial membrane 40 homolog, like |
| chr8_-_30791089 | 0.26 |
ENSDART00000147332
|
gstt1a
|
glutathione S-transferase theta 1a |
| chr12_+_10163585 | 0.26 |
ENSDART00000106191
|
psmc5
|
proteasome 26S subunit, ATPase 5 |
| chr8_+_471342 | 0.25 |
ENSDART00000167205
|
nudt12
|
nudix (nucleoside diphosphate linked moiety X)-type motif 12 |
| chr5_-_4493502 | 0.25 |
ENSDART00000067593
ENSDART00000190221 |
zgc:153704
|
zgc:153704 |
| chr7_+_23875269 | 0.24 |
ENSDART00000101406
|
rab39bb
|
RAB39B, member RAS oncogene family b |
| chr21_+_6961635 | 0.24 |
ENSDART00000137281
|
si:ch211-93g21.2
|
si:ch211-93g21.2 |
| chr5_+_23256187 | 0.24 |
ENSDART00000168717
ENSDART00000142915 |
si:dkey-125i10.3
|
si:dkey-125i10.3 |
| chr6_+_28205284 | 0.23 |
ENSDART00000160707
ENSDART00000190509 |
smx5
LSM2 (1 of many)
|
smx5 si:ch73-14h10.2 |
| chr9_+_41024973 | 0.23 |
ENSDART00000014660
ENSDART00000144467 |
ormdl1
|
ORMDL sphingolipid biosynthesis regulator 1 |
| chr2_+_47582681 | 0.23 |
ENSDART00000187579
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr13_-_32635859 | 0.22 |
ENSDART00000146249
ENSDART00000145395 ENSDART00000148040 ENSDART00000100650 |
matn3b
|
matrilin 3b |
| chr24_+_14713776 | 0.22 |
ENSDART00000134475
|
gdap1
|
ganglioside induced differentiation associated protein 1 |
| chr7_+_11543999 | 0.22 |
ENSDART00000173676
|
il16
|
interleukin 16 |
| chr16_-_43025885 | 0.22 |
ENSDART00000193146
ENSDART00000157302 |
si:dkey-7j14.5
|
si:dkey-7j14.5 |
| chr13_+_4205724 | 0.21 |
ENSDART00000134105
|
dlk2
|
delta-like 2 homolog (Drosophila) |
| chr21_-_4032650 | 0.21 |
ENSDART00000151648
|
ntng2b
|
netrin g2b |
| chr12_+_28854410 | 0.21 |
ENSDART00000152991
|
nfe2l1b
|
nuclear factor, erythroid 2-like 1b |
| chr23_+_4299887 | 0.21 |
ENSDART00000132604
|
l3mbtl1a
|
l(3)mbt-like 1a (Drosophila) |
| chr21_+_22423286 | 0.20 |
ENSDART00000133190
|
capslb
|
calcyphosine-like b |
| chr1_+_36651059 | 0.20 |
ENSDART00000187475
|
ednraa
|
endothelin receptor type Aa |
| chr8_+_68864 | 0.19 |
ENSDART00000164574
|
prr16
|
proline rich 16 |
| chr5_+_28973264 | 0.19 |
ENSDART00000005638
|
stxbp1b
|
syntaxin binding protein 1b |
| chr23_+_43255328 | 0.18 |
ENSDART00000102712
|
tgm2a
|
transglutaminase 2, C polypeptide A |
| chr21_-_4244514 | 0.18 |
ENSDART00000065880
|
uck1
|
uridine-cytidine kinase 1 |
| chr16_+_24681177 | 0.18 |
ENSDART00000058956
ENSDART00000189335 |
ywhabl
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide like |
| chr5_+_28972935 | 0.18 |
ENSDART00000193274
|
stxbp1b
|
syntaxin binding protein 1b |
| chr7_-_38638809 | 0.18 |
ENSDART00000144341
|
c6ast4
|
six-cysteine containing astacin protease 4 |
| chr16_+_34479064 | 0.18 |
ENSDART00000159965
|
si:ch211-255i3.4
|
si:ch211-255i3.4 |
| chr13_-_40726865 | 0.18 |
ENSDART00000099847
|
st3gal7
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 7 |
| chr8_-_18239494 | 0.18 |
ENSDART00000079989
ENSDART00000022959 |
guk1b
|
guanylate kinase 1b |
| chr20_-_26929685 | 0.17 |
ENSDART00000132556
|
ftr79
|
finTRIM family, member 79 |
| chr6_-_32314913 | 0.17 |
ENSDART00000051779
|
atg4c
|
autophagy related 4C, cysteine peptidase |
| chr11_-_21014572 | 0.17 |
ENSDART00000042220
|
slc6a14
|
solute carrier family 6 (amino acid transporter), member 14 |
| chr7_-_5316901 | 0.16 |
ENSDART00000181505
ENSDART00000124367 |
si:cabz01074946.1
|
si:cabz01074946.1 |
| chr1_+_20635190 | 0.16 |
ENSDART00000145418
ENSDART00000148518 ENSDART00000139461 ENSDART00000102969 ENSDART00000166479 |
spock3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr3_-_58798377 | 0.16 |
ENSDART00000161248
|
si:ch73-281f12.4
|
si:ch73-281f12.4 |
| chr15_+_42559910 | 0.16 |
ENSDART00000075785
|
cldn17
|
claudin 17 |
| chr8_+_23009662 | 0.16 |
ENSDART00000030885
|
uckl1a
|
uridine-cytidine kinase 1-like 1a |
| chr6_+_27146671 | 0.16 |
ENSDART00000156792
|
kif1aa
|
kinesin family member 1Aa |
| chr2_-_13216269 | 0.15 |
ENSDART00000149947
|
bcl2b
|
BCL2, apoptosis regulator b |
| chr19_+_9004256 | 0.15 |
ENSDART00000186401
ENSDART00000091443 |
si:ch211-81a5.1
|
si:ch211-81a5.1 |
| chr2_+_35854242 | 0.15 |
ENSDART00000134918
|
dhx9
|
DEAH (Asp-Glu-Ala-His) box helicase 9 |
| chr18_-_14691727 | 0.15 |
ENSDART00000010129
|
pdf
|
peptide deformylase, mitochondrial |
| chr5_-_6508250 | 0.15 |
ENSDART00000060535
|
crybb3
|
crystallin, beta B3 |
| chr2_+_47582488 | 0.15 |
ENSDART00000149967
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr9_-_23944470 | 0.15 |
ENSDART00000138754
|
col6a3
|
collagen, type VI, alpha 3 |
| chr1_+_44127292 | 0.14 |
ENSDART00000160542
|
cabp2a
|
calcium binding protein 2a |
| chr2_+_44545912 | 0.14 |
ENSDART00000155362
|
si:dkeyp-94h10.5
|
si:dkeyp-94h10.5 |
| chr2_-_32551178 | 0.14 |
ENSDART00000145603
|
smarcd3a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3a |
| chr3_-_32611833 | 0.14 |
ENSDART00000151282
|
si:ch73-248e21.5
|
si:ch73-248e21.5 |
| chr17_-_20897250 | 0.14 |
ENSDART00000088106
|
ank3b
|
ankyrin 3b |
| chr1_+_52518176 | 0.14 |
ENSDART00000003278
|
tacr3l
|
tachykinin receptor 3-like |
| chr7_+_57677120 | 0.14 |
ENSDART00000110623
|
arsj
|
arylsulfatase family, member J |
| chr7_-_32599669 | 0.14 |
ENSDART00000173752
|
kcna4
|
potassium voltage-gated channel, shaker-related subfamily, member 4 |
| chr8_-_23573084 | 0.14 |
ENSDART00000139084
|
wasb
|
Wiskott-Aldrich syndrome (eczema-thrombocytopenia) b |
| chr6_-_10964083 | 0.13 |
ENSDART00000181583
|
notum2
|
notum pectinacetylesterase 2 |
| chr3_+_55105066 | 0.13 |
ENSDART00000110848
|
si:ch211-5k11.8
|
si:ch211-5k11.8 |
| chr20_+_49119633 | 0.13 |
ENSDART00000151435
|
cd109
|
CD109 molecule |
| chr12_-_10275320 | 0.13 |
ENSDART00000170078
|
mpp2b
|
membrane protein, palmitoylated 2b (MAGUK p55 subfamily member 2) |
| chr5_+_33281105 | 0.13 |
ENSDART00000011337
|
surf1
|
surfeit 1 |
| chr21_-_40083432 | 0.12 |
ENSDART00000141160
ENSDART00000191195 |
slc13a5a
|
info solute carrier family 13 (sodium-dependent citrate transporter), member 5a |
| chr25_-_13676682 | 0.12 |
ENSDART00000090226
|
znf319b
|
zinc finger protein 319b |
| chr25_+_7671640 | 0.12 |
ENSDART00000145367
|
kcnj11l
|
potassium inwardly-rectifying channel, subfamily J, member 11, like |
| chr11_+_38332419 | 0.12 |
ENSDART00000005864
|
CDK18
|
si:dkey-166c18.1 |
| chr22_+_21972824 | 0.12 |
ENSDART00000183708
ENSDART00000133739 |
gna15.3
|
guanine nucleotide binding protein (G protein), alpha 15 (Gq class), tandem duplicate 3 |
| chr4_-_20118468 | 0.12 |
ENSDART00000078587
|
si:dkey-159a18.1
|
si:dkey-159a18.1 |
| chr20_-_17259573 | 0.12 |
ENSDART00000166397
|
AL928808.1
|
|
| chr16_-_38001040 | 0.12 |
ENSDART00000133861
ENSDART00000138711 ENSDART00000143846 ENSDART00000146564 |
si:ch211-198c19.3
|
si:ch211-198c19.3 |
| chr5_+_21931124 | 0.12 |
ENSDART00000137627
|
si:ch73-92i20.1
|
si:ch73-92i20.1 |
| chr6_-_13200585 | 0.11 |
ENSDART00000185321
|
eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr1_-_49544601 | 0.11 |
ENSDART00000145960
|
slc47a2.2
|
solute carrier family 47 (multidrug and toxin extrusion), member 2.2 |
| chr3_+_38793499 | 0.11 |
ENSDART00000083394
|
kcnj19a
|
potassium voltage-gated channel subfamily J member 19a |
| chr15_+_5321612 | 0.11 |
ENSDART00000174345
|
or113-1
|
odorant receptor, family A, subfamily 113, member 1 |
| chr4_+_25607743 | 0.11 |
ENSDART00000028297
|
acot14
|
acyl-CoA thioesterase 14 |
| chr1_+_55239710 | 0.10 |
ENSDART00000174846
|
si:ch211-286b5.2
|
si:ch211-286b5.2 |
| chr25_-_6557854 | 0.10 |
ENSDART00000181740
|
cspg4
|
chondroitin sulfate proteoglycan 4 |
| chr6_-_10912424 | 0.10 |
ENSDART00000036456
|
cycsb
|
cytochrome c, somatic b |
| chr18_-_29961784 | 0.10 |
ENSDART00000135601
ENSDART00000140661 |
si:ch73-103b9.2
|
si:ch73-103b9.2 |
| chr22_+_17192767 | 0.10 |
ENSDART00000130810
ENSDART00000183006 |
atpaf1
|
ATP synthase mitochondrial F1 complex assembly factor 1 |
| chr9_-_32730487 | 0.10 |
ENSDART00000147795
|
olig1
|
oligodendrocyte transcription factor 1 |
| chr17_+_24006792 | 0.10 |
ENSDART00000122415
|
si:ch211-63b16.4
|
si:ch211-63b16.4 |
| chr2_-_4797512 | 0.10 |
ENSDART00000160765
|
tnk2b
|
tyrosine kinase, non-receptor, 2b |
| chr22_+_35516440 | 0.10 |
ENSDART00000184191
|
hykk.2
|
hydroxylysine kinase, tandem duplicate 2 |
| chr2_-_36932253 | 0.10 |
ENSDART00000124217
|
map1sb
|
microtubule-associated protein 1Sb |
| chr23_+_44644911 | 0.09 |
ENSDART00000140799
|
zgc:85858
|
zgc:85858 |
| chr25_-_35996141 | 0.09 |
ENSDART00000149074
|
sall1b
|
spalt-like transcription factor 1b |
| chr20_-_5267600 | 0.09 |
ENSDART00000099258
|
cyp46a1.4
|
cytochrome P450, family 46, subfamily A, polypeptide 1, tandem duplicate 4 |
| chr15_+_5973909 | 0.09 |
ENSDART00000126886
ENSDART00000189618 |
igsf5b
|
immunoglobulin superfamily, member 5b |
| chr25_-_36286186 | 0.09 |
ENSDART00000182398
|
slc7a10b
|
solute carrier family 7 (neutral amino acid transporter light chain, asc system), member 10b |
| chr16_-_19349318 | 0.09 |
ENSDART00000020821
|
dnah11
|
dynein, axonemal, heavy chain 11 |
| chr5_+_69868911 | 0.09 |
ENSDART00000014649
ENSDART00000188215 ENSDART00000167385 |
ugt2a5
|
UDP glucuronosyltransferase 2 family, polypeptide A5 |
| chr6_+_612330 | 0.09 |
ENSDART00000166872
ENSDART00000191758 |
kynu
|
kynureninase |
| chr2_-_6039757 | 0.09 |
ENSDART00000013079
|
scp2a
|
sterol carrier protein 2a |
| chr16_+_40508882 | 0.09 |
ENSDART00000126129
|
nagpa
|
N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase |
| chr6_-_10963887 | 0.08 |
ENSDART00000151102
|
notum2
|
notum pectinacetylesterase 2 |
| chr10_+_3875716 | 0.08 |
ENSDART00000189268
ENSDART00000180624 |
TTC28
|
tetratricopeptide repeat domain 28 |
| chr2_-_11504347 | 0.08 |
ENSDART00000019392
|
sdr16c5a
|
short chain dehydrogenase/reductase family 16C, member 5a |
| chr25_+_25438322 | 0.08 |
ENSDART00000150364
|
LRRC56
|
si:ch211-103e16.5 |
| chr23_+_19606291 | 0.08 |
ENSDART00000139415
|
flnb
|
filamin B, beta (actin binding protein 278) |
| chr17_-_51938663 | 0.08 |
ENSDART00000179784
|
ERG28
|
ergosterol biosynthesis 28 homolog |
| chr4_-_69917450 | 0.08 |
ENSDART00000168915
|
znf1076
|
zinc finger protein 1076 |
| chr18_-_34156765 | 0.08 |
ENSDART00000059400
|
c18h3orf33
|
c18h3orf33 homolog (H. sapiens) |
| chr14_-_33360344 | 0.07 |
ENSDART00000181291
|
pimr117
|
Pim proto-oncogene, serine/threonine kinase, related 117 |
| chr11_-_40101246 | 0.07 |
ENSDART00000161083
|
tnfrsf9b
|
tumor necrosis factor receptor superfamily, member 9b |
| chr22_+_10781894 | 0.07 |
ENSDART00000081183
|
enc3
|
ectodermal-neural cortex 3 |
| chr3_+_32099507 | 0.07 |
ENSDART00000044238
|
zgc:92066
|
zgc:92066 |
| chr14_-_28568107 | 0.07 |
ENSDART00000042850
ENSDART00000145502 |
insb
|
preproinsulin b |
| chr12_+_9761685 | 0.07 |
ENSDART00000189522
|
ppp1r9bb
|
protein phosphatase 1, regulatory subunit 9Bb |
| chr3_+_27786601 | 0.07 |
ENSDART00000086994
|
nat15
|
N-acetyltransferase 15 (GCN5-related, putative) |
| chr7_-_18656069 | 0.07 |
ENSDART00000021559
|
coro1b
|
coronin, actin binding protein, 1B |
| chr22_+_1887750 | 0.07 |
ENSDART00000180917
|
si:dkey-15h8.17
|
si:dkey-15h8.17 |
| chr17_-_20897407 | 0.06 |
ENSDART00000149481
|
ank3b
|
ankyrin 3b |
| chr13_-_44738574 | 0.06 |
ENSDART00000074761
|
zfand3
|
zinc finger, AN1-type domain 3 |
| chr15_-_24178893 | 0.06 |
ENSDART00000077980
|
pipox
|
pipecolic acid oxidase |
| chr7_+_39624728 | 0.06 |
ENSDART00000173847
ENSDART00000173845 |
ptpn5
|
protein tyrosine phosphatase, non-receptor type 5 |
| chr6_-_10776254 | 0.06 |
ENSDART00000184008
|
gpr155b
|
G protein-coupled receptor 155b |
| chr10_+_20630444 | 0.06 |
ENSDART00000191182
|
prnpb
|
prion protein b |
| chr22_-_8725768 | 0.06 |
ENSDART00000189873
ENSDART00000181819 |
si:ch73-27e22.1
si:ch73-27e22.8
|
si:ch73-27e22.1 si:ch73-27e22.8 |
| chr4_-_29753895 | 0.06 |
ENSDART00000150742
|
si:dkey-191j3.1
|
si:dkey-191j3.1 |
| chr7_+_22702437 | 0.06 |
ENSDART00000182054
|
si:dkey-165a24.9
|
si:dkey-165a24.9 |
| chr7_+_65272470 | 0.06 |
ENSDART00000162471
|
bco1
|
beta-carotene oxygenase 1 |
| chr3_-_2589704 | 0.06 |
ENSDART00000187239
|
si:dkey-217f16.5
|
si:dkey-217f16.5 |
| chr12_-_17199381 | 0.06 |
ENSDART00000193292
|
lipf
|
lipase, gastric |
| chr24_+_2961098 | 0.06 |
ENSDART00000163760
ENSDART00000170835 |
eci2
|
enoyl-CoA delta isomerase 2 |
| chr25_-_3759322 | 0.06 |
ENSDART00000088077
|
zgc:158398
|
zgc:158398 |
| chr10_+_1899595 | 0.06 |
ENSDART00000040271
|
CU861476.1
|
|
| chr5_-_22621417 | 0.05 |
ENSDART00000138716
|
zgc:113208
|
zgc:113208 |
| chr21_+_21339311 | 0.05 |
ENSDART00000137693
ENSDART00000161820 |
sdhaf1
si:ch211-191j22.3
|
succinate dehydrogenase complex assembly factor 1 si:ch211-191j22.3 |
| chr14_-_26400501 | 0.05 |
ENSDART00000054189
|
pimr212
|
Pim proto-oncogene, serine/threonine kinase, related 212 |
| chr1_+_15226268 | 0.05 |
ENSDART00000109911
|
hgsnat
|
heparan-alpha-glucosaminide N-acetyltransferase |
| chr12_-_11649690 | 0.05 |
ENSDART00000149713
|
btbd16
|
BTB (POZ) domain containing 16 |
| chr23_+_14590767 | 0.05 |
ENSDART00000143675
|
si:rp71-79p20.2
|
si:rp71-79p20.2 |
| chr17_+_53284040 | 0.05 |
ENSDART00000165368
|
atxn3
|
ataxin 3 |
| chr25_+_10947743 | 0.05 |
ENSDART00000073381
|
mhc1lfa
|
major histocompatibility complex class I LFA |
| chr4_-_42391176 | 0.05 |
ENSDART00000189739
|
si:ch211-59d8.2
|
si:ch211-59d8.2 |
| chr6_+_14980761 | 0.05 |
ENSDART00000087782
|
mrps9
|
mitochondrial ribosomal protein S9 |
| chr24_-_25186895 | 0.05 |
ENSDART00000081029
|
plcxd2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of irx2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0097435 | fibril organization(GO:0097435) |
| 0.1 | 0.4 | GO:0014809 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 0.5 | GO:0019401 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) short-chain fatty acid catabolic process(GO:0019626) response to dexamethasone(GO:0071548) |
| 0.1 | 0.6 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.1 | 0.3 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.1 | 0.4 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.1 | 0.3 | GO:0072526 | pyridine nucleotide catabolic process(GO:0019364) NAD catabolic process(GO:0019677) pyridine-containing compound catabolic process(GO:0072526) |
| 0.1 | 0.2 | GO:0090155 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.1 | 0.3 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 0.3 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.1 | 0.3 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.2 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.1 | 0.5 | GO:0032096 | negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.0 | 0.4 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.0 | 0.3 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.3 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.5 | GO:0048385 | regulation of retinoic acid receptor signaling pathway(GO:0048385) |
| 0.0 | 0.2 | GO:0035176 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.0 | 0.1 | GO:0042940 | D-amino acid transport(GO:0042940) D-alanine transport(GO:0042941) D-serine transport(GO:0042942) |
| 0.0 | 0.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.4 | GO:0006596 | polyamine biosynthetic process(GO:0006596) |
| 0.0 | 0.1 | GO:0097052 | L-kynurenine metabolic process(GO:0097052) L-kynurenine catabolic process(GO:0097053) |
| 0.0 | 0.2 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.3 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.3 | GO:0007035 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.0 | 0.6 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.6 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.1 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.0 | 0.3 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.0 | 0.5 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.1 | GO:0032370 | positive regulation of lipid transport(GO:0032370) |
| 0.0 | 0.7 | GO:1903670 | regulation of sprouting angiogenesis(GO:1903670) |
| 0.0 | 0.1 | GO:0006337 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.1 | GO:0019477 | lysine catabolic process(GO:0006554) L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.0 | 0.5 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.2 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.5 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.4 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.1 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.0 | 0.1 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.2 | GO:0050926 | regulation of positive chemotaxis(GO:0050926) positive regulation of positive chemotaxis(GO:0050927) induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.1 | GO:0034553 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.3 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.1 | 0.4 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 0.2 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.2 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.3 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.3 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 1.0 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.3 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.1 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.3 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 1.0 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.2 | 0.5 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.1 | 0.3 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.4 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 0.4 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.1 | 0.4 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.1 | 0.6 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 0.3 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 0.2 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.1 | 0.2 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.1 | 0.3 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.2 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.3 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.1 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.0 | 0.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.2 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.2 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 0.3 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.9 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.6 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.4 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.3 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.1 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.3 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 0.5 | GO:0004675 | transmembrane receptor protein serine/threonine kinase activity(GO:0004675) |
| 0.0 | 0.1 | GO:0019202 | amino acid kinase activity(GO:0019202) |
| 0.0 | 0.1 | GO:0033781 | cholesterol 24-hydroxylase activity(GO:0033781) |
| 0.0 | 0.1 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.4 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.0 | 0.5 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.1 | GO:0031420 | potassium ion binding(GO:0030955) alkali metal ion binding(GO:0031420) |
| 0.0 | 0.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.3 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.1 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.2 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |