Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
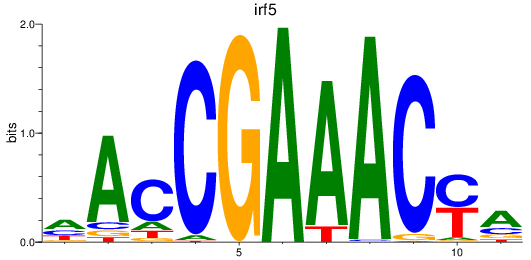
Results for irf5
Z-value: 2.58
Transcription factors associated with irf5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
irf5
|
ENSDARG00000045681 | interferon regulatory factor 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| irf5 | dr11_v1_chr4_-_13614797_13614936 | 0.85 | 3.6e-03 | Click! |
Activity profile of irf5 motif
Sorted Z-values of irf5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_69187585 | 5.26 |
ENSDART00000160499
ENSDART00000166258 |
marveld3
|
MARVEL domain containing 3 |
| chr20_-_43743700 | 3.53 |
ENSDART00000100620
|
si:dkeyp-50f7.2
|
si:dkeyp-50f7.2 |
| chr15_+_44201056 | 3.40 |
ENSDART00000162433
ENSDART00000148336 |
CU655961.4
|
|
| chr5_-_30382925 | 3.36 |
ENSDART00000125381
|
gig2o
|
grass carp reovirus (GCRV)-induced gene 2o |
| chr22_+_24623936 | 3.30 |
ENSDART00000160924
|
mcoln2
|
mucolipin 2 |
| chr7_+_46020508 | 3.03 |
ENSDART00000170294
|
ccne1
|
cyclin E1 |
| chr15_-_29162193 | 2.85 |
ENSDART00000138449
ENSDART00000099885 |
xaf1
|
XIAP associated factor 1 |
| chr15_-_33964897 | 2.83 |
ENSDART00000172075
ENSDART00000158126 ENSDART00000160456 |
lsr
|
lipolysis stimulated lipoprotein receptor |
| chr17_+_22381215 | 2.82 |
ENSDART00000162670
ENSDART00000128875 |
slc8a1b
|
solute carrier family 8 (sodium/calcium exchanger), member 1b |
| chr18_+_44532668 | 2.78 |
ENSDART00000140672
|
st14a
|
suppression of tumorigenicity 14 (colon carcinoma) a |
| chr14_+_32918484 | 2.68 |
ENSDART00000105721
|
lnx2b
|
ligand of numb-protein X 2b |
| chr15_+_25452092 | 2.62 |
ENSDART00000009545
|
pak4
|
p21 protein (Cdc42/Rac)-activated kinase 4 |
| chr14_-_41467497 | 2.59 |
ENSDART00000181220
|
mid1ip1l
|
MID1 interacting protein 1, like |
| chr13_-_37631092 | 2.51 |
ENSDART00000108855
|
si:dkey-188i13.7
|
si:dkey-188i13.7 |
| chr18_+_44532199 | 2.50 |
ENSDART00000135386
|
st14a
|
suppression of tumorigenicity 14 (colon carcinoma) a |
| chr13_-_23051766 | 2.49 |
ENSDART00000111774
|
supv3l1
|
SUV3-like helicase |
| chr18_+_44532370 | 2.45 |
ENSDART00000086952
|
st14a
|
suppression of tumorigenicity 14 (colon carcinoma) a |
| chr21_+_21010786 | 2.36 |
ENSDART00000079692
|
ndr1
|
nodal-related 1 |
| chr25_+_28862660 | 2.36 |
ENSDART00000154681
|
si:ch211-106e7.2
|
si:ch211-106e7.2 |
| chr5_+_57442271 | 2.32 |
ENSDART00000097395
|
prpf4
|
PRP4 pre-mRNA processing factor 4 homolog (yeast) |
| chr20_+_6535438 | 2.29 |
ENSDART00000145763
|
si:ch211-191a24.4
|
si:ch211-191a24.4 |
| chr25_+_15997957 | 2.26 |
ENSDART00000140047
|
ppfibp2b
|
PTPRF interacting protein, binding protein 2b (liprin beta 2) |
| chr24_-_34680956 | 2.26 |
ENSDART00000171009
|
ctnna1
|
catenin (cadherin-associated protein), alpha 1 |
| chr5_+_57726425 | 2.25 |
ENSDART00000134684
|
fdxacb1
|
ferredoxin-fold anticodon binding domain containing 1 |
| chr20_+_6535176 | 2.25 |
ENSDART00000054652
|
si:ch211-191a24.4
|
si:ch211-191a24.4 |
| chr15_+_44184367 | 2.25 |
ENSDART00000162918
ENSDART00000110060 |
zgc:165514
|
zgc:165514 |
| chr19_+_42227400 | 2.22 |
ENSDART00000131574
ENSDART00000135436 |
jtb
|
jumping translocation breakpoint |
| chr14_-_33278084 | 2.22 |
ENSDART00000132850
|
stard14
|
START domain containing 14 |
| chr17_+_30894431 | 2.21 |
ENSDART00000127996
|
degs2
|
delta(4)-desaturase, sphingolipid 2 |
| chr1_-_55750208 | 2.12 |
ENSDART00000142244
|
dnajb1b
|
DnaJ (Hsp40) homolog, subfamily B, member 1b |
| chr11_-_40257225 | 2.09 |
ENSDART00000139009
|
TRIM62 (1 of many)
|
si:ch211-193i15.2 |
| chr7_-_9717020 | 2.01 |
ENSDART00000029668
|
lrrk1
|
leucine-rich repeat kinase 1 |
| chr9_+_41459759 | 2.01 |
ENSDART00000132501
ENSDART00000100265 |
nemp2
|
nuclear envelope integral membrane protein 2 |
| chr2_-_21438492 | 1.97 |
ENSDART00000046098
|
plcd1b
|
phospholipase C, delta 1b |
| chr23_+_2666944 | 1.97 |
ENSDART00000192861
|
CABZ01057928.1
|
|
| chr3_-_36127234 | 1.93 |
ENSDART00000130917
|
coil
|
coilin p80 |
| chr25_+_16646113 | 1.89 |
ENSDART00000110426
|
cecr2
|
cat eye syndrome chromosome region, candidate 2 |
| chr18_+_44532883 | 1.89 |
ENSDART00000121994
|
st14a
|
suppression of tumorigenicity 14 (colon carcinoma) a |
| chr10_-_15879569 | 1.87 |
ENSDART00000136789
|
tjp2a
|
tight junction protein 2a (zona occludens 2) |
| chr23_-_10745288 | 1.87 |
ENSDART00000140745
ENSDART00000013768 |
eif4e3
|
eukaryotic translation initiation factor 4E family member 3 |
| chr5_-_29531948 | 1.86 |
ENSDART00000098360
|
arrdc1a
|
arrestin domain containing 1a |
| chr19_+_4968947 | 1.85 |
ENSDART00000003634
ENSDART00000134808 |
stard3
|
StAR-related lipid transfer (START) domain containing 3 |
| chr22_-_17688868 | 1.82 |
ENSDART00000012336
ENSDART00000147070 |
tjp3
|
tight junction protein 3 |
| chr25_+_20272145 | 1.82 |
ENSDART00000109605
|
si:dkey-219c3.2
|
si:dkey-219c3.2 |
| chr7_+_19600262 | 1.79 |
ENSDART00000007310
|
zgc:171731
|
zgc:171731 |
| chr14_-_33277743 | 1.78 |
ENSDART00000048130
|
stard14
|
START domain containing 14 |
| chr24_-_26885897 | 1.76 |
ENSDART00000180512
|
fndc3bb
|
fibronectin type III domain containing 3Bb |
| chr3_+_32571929 | 1.75 |
ENSDART00000151025
|
si:ch73-248e21.1
|
si:ch73-248e21.1 |
| chr19_-_15420678 | 1.75 |
ENSDART00000151454
ENSDART00000027697 |
serinc2
|
serine incorporator 2 |
| chr9_+_3055566 | 1.74 |
ENSDART00000189906
ENSDART00000175891 ENSDART00000093021 |
ppp1r9ala
|
protein phosphatase 1 regulatory subunit 9A-like A |
| chr5_-_67241633 | 1.70 |
ENSDART00000114783
|
clip1a
|
CAP-GLY domain containing linker protein 1a |
| chr16_+_19637384 | 1.69 |
ENSDART00000184773
ENSDART00000191895 ENSDART00000182020 ENSDART00000135359 |
macc1
|
metastasis associated in colon cancer 1 |
| chr5_+_58455488 | 1.67 |
ENSDART00000038602
ENSDART00000127958 |
slc37a2
|
solute carrier family 37 (glucose-6-phosphate transporter), member 2 |
| chr12_+_17154655 | 1.65 |
ENSDART00000028003
|
ankrd22
|
ankyrin repeat domain 22 |
| chr13_-_33700461 | 1.64 |
ENSDART00000160520
|
mad2l1bp
|
MAD2L1 binding protein |
| chr15_+_31911989 | 1.58 |
ENSDART00000111472
|
brca2
|
breast cancer 2, early onset |
| chr5_-_12587053 | 1.56 |
ENSDART00000162780
|
vsig10
|
V-set and immunoglobulin domain containing 10 |
| chr15_-_17071328 | 1.54 |
ENSDART00000122617
|
si:ch211-24o10.6
|
si:ch211-24o10.6 |
| chr6_-_1566407 | 1.53 |
ENSDART00000112118
|
trim107
|
tripartite motif containing 107 |
| chr16_-_41667101 | 1.52 |
ENSDART00000084528
|
atp2c1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr11_+_39969048 | 1.52 |
ENSDART00000193693
|
per3
|
period circadian clock 3 |
| chr25_-_27665978 | 1.52 |
ENSDART00000123590
|
si:ch211-91p5.3
|
si:ch211-91p5.3 |
| chr9_+_23765587 | 1.50 |
ENSDART00000145120
|
si:ch211-219a4.3
|
si:ch211-219a4.3 |
| chr18_+_45666489 | 1.50 |
ENSDART00000180147
ENSDART00000151351 |
prrg4
|
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) |
| chr5_+_39099172 | 1.49 |
ENSDART00000006079
|
bmp2k
|
BMP2 inducible kinase |
| chr21_+_20903244 | 1.49 |
ENSDART00000186193
|
c7b
|
complement component 7b |
| chr10_-_22150419 | 1.49 |
ENSDART00000006173
|
cldn7b
|
claudin 7b |
| chr23_+_31942040 | 1.48 |
ENSDART00000088607
|
nemp1
|
nuclear envelope integral membrane protein 1 |
| chr5_+_50913357 | 1.47 |
ENSDART00000092938
|
col4a3bpa
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein a |
| chr8_-_49304602 | 1.46 |
ENSDART00000147020
|
prickle3
|
prickle homolog 3 |
| chr15_-_1835189 | 1.46 |
ENSDART00000154369
|
taf15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr6_-_1566581 | 1.46 |
ENSDART00000192993
|
trim107
|
tripartite motif containing 107 |
| chr16_-_26525901 | 1.43 |
ENSDART00000110260
|
l3mbtl1b
|
l(3)mbt-like 1b (Drosophila) |
| chr20_+_20751425 | 1.43 |
ENSDART00000177048
|
ppp1r13bb
|
protein phosphatase 1, regulatory subunit 13Bb |
| chr25_-_12809361 | 1.42 |
ENSDART00000162750
|
ca5a
|
carbonic anhydrase Va |
| chr3_+_36127287 | 1.40 |
ENSDART00000058605
ENSDART00000182500 |
scpep1
|
serine carboxypeptidase 1 |
| chr20_+_1316803 | 1.36 |
ENSDART00000152586
ENSDART00000152165 |
nup43
|
nucleoporin 43 |
| chr10_+_35152928 | 1.36 |
ENSDART00000063418
|
nsun5
|
NOP2/Sun domain family, member 5 |
| chr8_-_19266325 | 1.35 |
ENSDART00000036148
ENSDART00000137994 |
zgc:77486
|
zgc:77486 |
| chr16_+_27383717 | 1.35 |
ENSDART00000132329
ENSDART00000136256 |
stx17
|
syntaxin 17 |
| chr17_-_24879003 | 1.34 |
ENSDART00000123147
|
zbtb8a
|
zinc finger and BTB domain containing 8A |
| chr12_+_24060894 | 1.34 |
ENSDART00000021298
|
asb3
|
ankyrin repeat and SOCS box containing 3 |
| chr21_-_22122312 | 1.33 |
ENSDART00000101726
|
slc35f2
|
solute carrier family 35, member F2 |
| chr2_+_34967022 | 1.32 |
ENSDART00000134926
|
astn1
|
astrotactin 1 |
| chr19_+_9113932 | 1.32 |
ENSDART00000060442
|
setdb1a
|
SET domain, bifurcated 1a |
| chr18_+_37015185 | 1.32 |
ENSDART00000191305
|
sipa1l3
|
signal-induced proliferation-associated 1 like 3 |
| chr25_+_16080181 | 1.32 |
ENSDART00000061753
|
far1
|
fatty acyl CoA reductase 1 |
| chr9_+_17862858 | 1.30 |
ENSDART00000166566
|
dgkh
|
diacylglycerol kinase, eta |
| chr9_+_32178374 | 1.30 |
ENSDART00000078576
|
coq10b
|
coenzyme Q10B |
| chr20_+_13783040 | 1.29 |
ENSDART00000115329
ENSDART00000152497 |
lpgat1
|
lysophosphatidylglycerol acyltransferase 1 |
| chr22_+_32235922 | 1.29 |
ENSDART00000114531
ENSDART00000136125 |
rbm15b
|
RNA binding motif protein 15B |
| chr14_-_31814149 | 1.29 |
ENSDART00000173393
|
arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chr5_+_39099380 | 1.27 |
ENSDART00000166657
|
bmp2k
|
BMP2 inducible kinase |
| chr10_+_4046448 | 1.26 |
ENSDART00000123086
ENSDART00000052268 |
pitpnb
|
phosphatidylinositol transfer protein, beta |
| chr23_-_29553430 | 1.26 |
ENSDART00000157773
ENSDART00000126384 |
ube4b
|
ubiquitination factor E4B, UFD2 homolog (S. cerevisiae) |
| chr25_+_16079913 | 1.26 |
ENSDART00000146350
|
far1
|
fatty acyl CoA reductase 1 |
| chr15_-_1484795 | 1.25 |
ENSDART00000129356
|
si:dkeyp-97b10.3
|
si:dkeyp-97b10.3 |
| chr5_+_20030414 | 1.24 |
ENSDART00000181430
ENSDART00000047841 ENSDART00000182813 |
sgsm1a
|
small G protein signaling modulator 1a |
| chr2_-_37353098 | 1.24 |
ENSDART00000056522
|
skila
|
SKI-like proto-oncogene a |
| chr13_+_25364324 | 1.23 |
ENSDART00000187471
|
chuk
|
conserved helix-loop-helix ubiquitous kinase |
| chr4_+_40018440 | 1.23 |
ENSDART00000110827
|
si:dkey-199k11.6
|
si:dkey-199k11.6 |
| chr16_+_26601364 | 1.22 |
ENSDART00000087537
|
epb41l4b
|
erythrocyte membrane protein band 4.1 like 4B |
| chr6_-_1566186 | 1.22 |
ENSDART00000156305
|
trim107
|
tripartite motif containing 107 |
| chr6_+_58280936 | 1.22 |
ENSDART00000155244
|
ralgapb
|
Ral GTPase activating protein, beta subunit (non-catalytic) |
| chr17_-_2039511 | 1.22 |
ENSDART00000160223
|
spint1a
|
serine peptidase inhibitor, Kunitz type 1 a |
| chr20_+_1316495 | 1.21 |
ENSDART00000064439
|
nup43
|
nucleoporin 43 |
| chr22_+_18319666 | 1.19 |
ENSDART00000033103
|
gatad2ab
|
GATA zinc finger domain containing 2Ab |
| chr21_+_19319804 | 1.19 |
ENSDART00000063621
|
abraxas2a
|
abraxas 2a, BRISC complex subunit |
| chr14_+_15768942 | 1.17 |
ENSDART00000158998
|
ergic1
|
endoplasmic reticulum-golgi intermediate compartment 1 |
| chr4_-_20222182 | 1.16 |
ENSDART00000132464
|
gsap
|
gamma-secretase activating protein |
| chr6_+_44163727 | 1.13 |
ENSDART00000064878
|
gxylt2
|
glucoside xylosyltransferase 2 |
| chr18_+_2153530 | 1.13 |
ENSDART00000158255
|
pygo1
|
pygopus family PHD finger 1 |
| chr12_-_28349026 | 1.13 |
ENSDART00000183768
ENSDART00000152998 |
zgc:195081
|
zgc:195081 |
| chr2_+_38025260 | 1.12 |
ENSDART00000075905
|
hnrnpc
|
heterogeneous nuclear ribonucleoprotein C |
| chr6_+_49771626 | 1.12 |
ENSDART00000134207
|
ctsz
|
cathepsin Z |
| chr19_-_10214264 | 1.10 |
ENSDART00000053300
ENSDART00000148225 |
znf865
|
zinc finger protein 865 |
| chr6_+_48348415 | 1.09 |
ENSDART00000064826
|
mov10a
|
Mov10 RISC complex RNA helicase a |
| chr20_-_6476705 | 1.08 |
ENSDART00000077095
|
trappc8
|
trafficking protein particle complex 8 |
| chr12_-_41759686 | 1.06 |
ENSDART00000172175
ENSDART00000165152 |
ppp2r2d
|
protein phosphatase 2, regulatory subunit B, delta |
| chr6_-_9646275 | 1.06 |
ENSDART00000012903
|
wdr12
|
WD repeat domain 12 |
| chr20_+_27087539 | 1.06 |
ENSDART00000062094
|
tmem251
|
transmembrane protein 251 |
| chr11_-_34151487 | 1.05 |
ENSDART00000173039
|
atp13a3
|
ATPase 13A3 |
| chr14_+_22076596 | 1.04 |
ENSDART00000106147
ENSDART00000100278 ENSDART00000131489 |
slc43a1a
|
solute carrier family 43 (amino acid system L transporter), member 1a |
| chr25_+_8447565 | 1.03 |
ENSDART00000142090
|
fanci
|
Fanconi anemia, complementation group I |
| chr7_-_6441865 | 1.02 |
ENSDART00000172831
|
hist1h2a10
|
histone cluster 1 H2A family member 10 |
| chr23_-_29553691 | 1.01 |
ENSDART00000053804
|
ube4b
|
ubiquitination factor E4B, UFD2 homolog (S. cerevisiae) |
| chr19_-_29303788 | 1.00 |
ENSDART00000112167
|
srfbp1
|
serum response factor binding protein 1 |
| chr12_-_3077395 | 0.98 |
ENSDART00000002867
ENSDART00000126315 |
rfng
|
RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr22_-_4439311 | 0.97 |
ENSDART00000169317
|
uhrf1
|
ubiquitin-like with PHD and ring finger domains 1 |
| chr12_-_16452200 | 0.96 |
ENSDART00000037601
|
rpp30
|
ribonuclease P/MRP 30 subunit |
| chr13_+_24263049 | 0.96 |
ENSDART00000135992
ENSDART00000088005 |
abcb10
|
ATP-binding cassette, sub-family B (MDR/TAP), member 10 |
| chr12_-_24060358 | 0.95 |
ENSDART00000050831
|
chac2
|
ChaC, cation transport regulator homolog 2 (E. coli) |
| chr2_+_34967210 | 0.95 |
ENSDART00000141796
|
astn1
|
astrotactin 1 |
| chr21_+_17301790 | 0.95 |
ENSDART00000145057
|
tsc1b
|
TSC complex subunit 1b |
| chr10_+_7719796 | 0.95 |
ENSDART00000191795
|
ggcx
|
gamma-glutamyl carboxylase |
| chr1_-_29747702 | 0.95 |
ENSDART00000133225
ENSDART00000189670 |
spp2
|
secreted phosphoprotein 2 |
| chr22_-_6499055 | 0.92 |
ENSDART00000142235
ENSDART00000183588 |
si:dkey-19a16.7
|
si:dkey-19a16.7 |
| chr9_-_23765480 | 0.91 |
ENSDART00000027212
|
si:ch211-219a4.6
|
si:ch211-219a4.6 |
| chr6_-_10728057 | 0.90 |
ENSDART00000002247
|
sp3b
|
Sp3b transcription factor |
| chr23_+_31000243 | 0.89 |
ENSDART00000085263
|
selenoi
|
selenoprotein I |
| chr8_+_18545933 | 0.89 |
ENSDART00000148806
|
tab3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chr11_+_2855430 | 0.88 |
ENSDART00000172837
|
kif21b
|
kinesin family member 21B |
| chr22_+_18319230 | 0.88 |
ENSDART00000184747
ENSDART00000184649 |
gatad2ab
|
GATA zinc finger domain containing 2Ab |
| chr7_-_20103384 | 0.87 |
ENSDART00000052902
|
acap1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr9_-_55772937 | 0.87 |
ENSDART00000159192
|
akap17a
|
A kinase (PRKA) anchor protein 17A |
| chr6_+_11250316 | 0.86 |
ENSDART00000137122
|
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr3_-_55650771 | 0.86 |
ENSDART00000162413
|
axin2
|
axin 2 (conductin, axil) |
| chr7_+_54222156 | 0.85 |
ENSDART00000165201
ENSDART00000158518 |
pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr1_-_1885516 | 0.84 |
ENSDART00000122187
ENSDART00000131675 |
si:ch211-132g1.3
|
si:ch211-132g1.3 |
| chr6_-_33916756 | 0.84 |
ENSDART00000137447
ENSDART00000138488 |
nasp
|
nuclear autoantigenic sperm protein (histone-binding) |
| chr15_+_714203 | 0.84 |
ENSDART00000153847
|
si:dkey-7i4.24
|
si:dkey-7i4.24 |
| chr1_+_38153944 | 0.84 |
ENSDART00000135666
|
galnt7
|
UDP-N-acetyl-alpha-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase 7 |
| chr6_-_23002373 | 0.83 |
ENSDART00000037709
ENSDART00000170369 |
nol11
|
nucleolar protein 11 |
| chr25_+_27410352 | 0.83 |
ENSDART00000154362
|
pot1
|
protection of telomeres 1 homolog |
| chr6_+_49771372 | 0.82 |
ENSDART00000063251
|
ctsz
|
cathepsin Z |
| chr8_+_18545539 | 0.81 |
ENSDART00000089274
|
tab3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chr17_+_52300018 | 0.81 |
ENSDART00000190302
|
esrrb
|
estrogen-related receptor beta |
| chr15_+_1534644 | 0.80 |
ENSDART00000130413
|
smc4
|
structural maintenance of chromosomes 4 |
| chr22_+_336256 | 0.80 |
ENSDART00000019155
|
btg2
|
B-cell translocation gene 2 |
| chr7_+_32693890 | 0.80 |
ENSDART00000121972
|
slc39a13
|
solute carrier family 39 (zinc transporter), member 13 |
| chr8_+_31008287 | 0.80 |
ENSDART00000129882
ENSDART00000180694 |
ptgesl
|
prostaglandin E synthase 2-like |
| chr3_-_55650417 | 0.79 |
ENSDART00000171441
|
axin2
|
axin 2 (conductin, axil) |
| chr14_-_49992709 | 0.78 |
ENSDART00000159988
|
fam193b
|
family with sequence similarity 193, member B |
| chr3_+_15773991 | 0.77 |
ENSDART00000089923
|
znf652
|
zinc finger protein 652 |
| chr23_-_35396845 | 0.76 |
ENSDART00000142038
ENSDART00000049373 ENSDART00000181978 ENSDART00000171357 |
cmtr1
|
cap methyltransferase 1 |
| chr7_+_38808027 | 0.75 |
ENSDART00000052323
|
harbi1
|
harbinger transposase derived 1 |
| chr16_-_32303835 | 0.75 |
ENSDART00000191408
|
mms22l
|
MMS22-like, DNA repair protein |
| chr6_-_45869127 | 0.75 |
ENSDART00000062459
ENSDART00000180563 |
rbm19
|
RNA binding motif protein 19 |
| chr22_-_11648094 | 0.73 |
ENSDART00000191791
|
dpp4
|
dipeptidyl-peptidase 4 |
| chr21_-_30273418 | 0.72 |
ENSDART00000187069
ENSDART00000181492 |
zgc:175066
|
zgc:175066 |
| chr8_+_20157798 | 0.71 |
ENSDART00000124809
|
acsbg2
|
acyl-CoA synthetase bubblegum family member 2 |
| chr8_+_23802384 | 0.71 |
ENSDART00000137820
|
si:ch211-163l21.8
|
si:ch211-163l21.8 |
| chr16_+_35595312 | 0.71 |
ENSDART00000170438
|
si:ch211-1i11.3
|
si:ch211-1i11.3 |
| chr21_+_33187992 | 0.71 |
ENSDART00000162745
ENSDART00000188388 |
BX072577.1
|
|
| chr20_+_3916406 | 0.71 |
ENSDART00000064365
|
si:ch73-111k22.2
|
si:ch73-111k22.2 |
| chr14_+_16345003 | 0.70 |
ENSDART00000003040
ENSDART00000165193 |
itln3
|
intelectin 3 |
| chr5_+_44805269 | 0.69 |
ENSDART00000136965
|
ctsla
|
cathepsin La |
| chr5_-_13086616 | 0.68 |
ENSDART00000051664
|
ypel1
|
yippee-like 1 |
| chr21_-_13055195 | 0.67 |
ENSDART00000133517
|
myorg
|
myogenesis regulating glycosidase (putative) |
| chr5_-_37820878 | 0.67 |
ENSDART00000165513
|
AL935065.1
|
Danio rerio uncharacterized LOC567472 (LOC567472), mRNA. |
| chr7_-_6444011 | 0.67 |
ENSDART00000173010
|
zgc:112234
|
zgc:112234 |
| chr3_-_23596809 | 0.67 |
ENSDART00000156897
|
ube2z
|
ubiquitin-conjugating enzyme E2Z |
| chr23_-_12906228 | 0.67 |
ENSDART00000138807
|
ndnl2
|
necdin-like 2 |
| chr11_-_20071642 | 0.67 |
ENSDART00000162931
ENSDART00000159928 ENSDART00000191443 ENSDART00000121722 |
si:dkey-274m17.3
|
si:dkey-274m17.3 |
| chr24_-_10828560 | 0.66 |
ENSDART00000132282
|
fam49bb
|
family with sequence similarity 49, member Bb |
| chr9_-_28274932 | 0.65 |
ENSDART00000137582
ENSDART00000146932 |
creb1b
|
cAMP responsive element binding protein 1b |
| chr18_+_45708744 | 0.65 |
ENSDART00000077341
|
depdc7
|
DEP domain containing 7 |
| chr9_+_23003208 | 0.64 |
ENSDART00000021060
|
eaf2
|
ELL associated factor 2 |
| chr10_-_2971407 | 0.62 |
ENSDART00000132526
|
marveld2a
|
MARVEL domain containing 2a |
| chr7_-_20865005 | 0.61 |
ENSDART00000190752
|
fis1
|
fission, mitochondrial 1 |
| chr24_+_8904741 | 0.60 |
ENSDART00000140924
|
gnal
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr4_-_20108833 | 0.59 |
ENSDART00000100867
|
fam3c
|
family with sequence similarity 3, member C |
| chr4_-_17669881 | 0.59 |
ENSDART00000066997
|
dram1
|
DNA-damage regulated autophagy modulator 1 |
| chr2_+_10007113 | 0.58 |
ENSDART00000155213
|
slc35a3b
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3b |
| chr7_+_8670398 | 0.57 |
ENSDART00000164195
|
si:ch211-1o7.2
|
si:ch211-1o7.2 |
| chr5_+_36895860 | 0.56 |
ENSDART00000134493
|
srsf7a
|
serine/arginine-rich splicing factor 7a |
| chr24_+_37688729 | 0.55 |
ENSDART00000137017
|
h3f3d
|
H3 histone, family 3D |
| chr11_-_3381343 | 0.55 |
ENSDART00000002545
|
mcrs1
|
microspherule protein 1 |
| chr12_-_27242498 | 0.55 |
ENSDART00000152609
ENSDART00000152170 |
si:dkey-11c5.11
|
si:dkey-11c5.11 |
| chr14_-_31060082 | 0.53 |
ENSDART00000111601
ENSDART00000161113 |
mbnl3
|
muscleblind-like splicing regulator 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of irf5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.3 | GO:0010633 | negative regulation of epithelial cell migration(GO:0010633) |
| 0.6 | 1.7 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.4 | 0.4 | GO:0008584 | male gonad development(GO:0008584) |
| 0.4 | 1.9 | GO:0030576 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.4 | 3.6 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.4 | 2.5 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.3 | 1.3 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.3 | 1.5 | GO:0055071 | manganese ion homeostasis(GO:0055071) |
| 0.3 | 1.7 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.3 | 0.8 | GO:1901836 | regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901836) |
| 0.3 | 1.3 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.2 | 1.5 | GO:0060876 | semicircular canal formation(GO:0060876) |
| 0.2 | 1.2 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.2 | 0.8 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.2 | 1.8 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.2 | 1.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.2 | 2.8 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.2 | 1.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.2 | 1.0 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.2 | 2.2 | GO:0034311 | diol metabolic process(GO:0034311) |
| 0.2 | 0.6 | GO:0035522 | monoubiquitinated protein deubiquitination(GO:0035520) monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 | 2.4 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.1 | 1.2 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.1 | 2.8 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 0.8 | GO:2000279 | negative regulation of DNA biosynthetic process(GO:2000279) |
| 0.1 | 1.4 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.1 | 1.9 | GO:0007530 | sex determination(GO:0007530) female gonad development(GO:0008585) |
| 0.1 | 2.3 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.1 | 1.3 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 1.2 | GO:0042987 | beta-amyloid formation(GO:0034205) amyloid precursor protein catabolic process(GO:0042987) |
| 0.1 | 0.8 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.1 | 1.1 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.1 | 1.6 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.1 | 0.6 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.1 | 1.0 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.1 | 1.1 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 0.5 | GO:0045124 | regulation of bone resorption(GO:0045124) |
| 0.1 | 1.3 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.1 | 0.6 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 0.3 | GO:0039529 | RIG-I signaling pathway(GO:0039529) regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039531) regulation of RIG-I signaling pathway(GO:0039535) |
| 0.1 | 1.6 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.1 | 1.9 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 0.8 | GO:0001516 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.1 | 0.9 | GO:0014028 | notochord formation(GO:0014028) |
| 0.1 | 0.4 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.1 | 0.7 | GO:0044090 | positive regulation of vacuole organization(GO:0044090) |
| 0.1 | 2.7 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.1 | 0.9 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 10.8 | GO:0008544 | epidermis development(GO:0008544) |
| 0.1 | 2.6 | GO:0048920 | posterior lateral line neuromast primordium migration(GO:0048920) |
| 0.1 | 0.5 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.1 | 0.9 | GO:0046849 | bone remodeling(GO:0046849) |
| 0.1 | 0.7 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.1 | 1.0 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.1 | 1.3 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone metabolic process(GO:1901661) quinone biosynthetic process(GO:1901663) |
| 0.1 | 2.1 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.1 | 1.3 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 2.5 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.1 | 3.8 | GO:0071166 | ribonucleoprotein complex localization(GO:0071166) |
| 0.1 | 1.1 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 0.8 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.1 | 3.0 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.1 | 1.2 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.1 | 0.5 | GO:0046685 | response to arsenic-containing substance(GO:0046685) |
| 0.1 | 0.5 | GO:2000290 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.1 | 1.3 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 1.5 | GO:0019835 | cytolysis(GO:0019835) |
| 0.1 | 1.8 | GO:2001236 | regulation of extrinsic apoptotic signaling pathway(GO:2001236) |
| 0.1 | 0.1 | GO:0030728 | ovulation(GO:0030728) |
| 0.1 | 1.0 | GO:0021654 | rhombomere boundary formation(GO:0021654) |
| 0.1 | 1.0 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 0.4 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.0 | 0.7 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.8 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 2.2 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 0.2 | GO:0048387 | ventricular trabecula myocardium morphogenesis(GO:0003222) negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 1.4 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.6 | GO:0014812 | muscle cell migration(GO:0014812) |
| 0.0 | 0.7 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 1.8 | GO:0030301 | cholesterol transport(GO:0030301) |
| 0.0 | 1.3 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.3 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.0 | 0.6 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 1.8 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 1.0 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.7 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 2.6 | GO:0002573 | myeloid leukocyte differentiation(GO:0002573) |
| 0.0 | 0.2 | GO:0048662 | regulation of smooth muscle cell proliferation(GO:0048660) negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.0 | 0.3 | GO:0006566 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.0 | 1.9 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 2.3 | GO:0001894 | tissue homeostasis(GO:0001894) |
| 0.0 | 1.7 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 2.3 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.4 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.2 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.2 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 1.2 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.7 | GO:0050870 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.0 | 0.1 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.1 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 1.0 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 2.0 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 1.6 | GO:0007492 | endoderm development(GO:0007492) |
| 0.0 | 1.6 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.4 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 2.5 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.3 | GO:0003374 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.4 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 1.0 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 1.2 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 0.3 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.4 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.0 | 0.5 | GO:0070972 | protein localization to endoplasmic reticulum(GO:0070972) |
| 0.0 | 0.1 | GO:0015961 | diadenosine polyphosphate metabolic process(GO:0015959) diadenosine polyphosphate catabolic process(GO:0015961) |
| 0.0 | 1.4 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.2 | GO:0051382 | kinetochore assembly(GO:0051382) negative regulation of cysteine-type endopeptidase activity(GO:2000117) |
| 0.0 | 0.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 1.3 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.4 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.6 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 1.8 | GO:0045786 | negative regulation of cell cycle(GO:0045786) |
| 0.0 | 1.0 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.0 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.4 | 1.1 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.4 | 1.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.3 | 2.5 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.3 | 1.9 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.2 | 1.0 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.2 | 2.6 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.2 | 1.8 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.2 | 1.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.2 | 1.0 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.2 | 1.3 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.2 | 1.9 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.2 | 0.6 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 0.7 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 2.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 2.1 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 1.9 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 2.3 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.1 | 1.5 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 1.8 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 0.6 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.1 | 0.6 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 3.5 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 0.6 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 0.2 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.1 | 1.1 | GO:0044545 | NSL complex(GO:0044545) |
| 0.1 | 8.5 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 0.7 | GO:0035101 | FACT complex(GO:0035101) |
| 0.1 | 1.7 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 1.0 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.1 | 2.1 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 1.8 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.4 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 1.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 2.2 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.8 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 1.0 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 1.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.5 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 2.8 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.2 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.6 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 2.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 3.4 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 1.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.3 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.5 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.2 | GO:0034451 | acrosomal vesicle(GO:0001669) centriolar satellite(GO:0034451) |
| 0.0 | 0.2 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.0 | 0.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.1 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.1 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 1.3 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 1.3 | GO:0016607 | nuclear speck(GO:0016607) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0016436 | rRNA (uridine) methyltransferase activity(GO:0016436) |
| 0.6 | 1.7 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.4 | 3.3 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.4 | 1.9 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.4 | 1.9 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.4 | 2.6 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.3 | 2.4 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.3 | 2.3 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.3 | 2.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.3 | 1.2 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.3 | 1.4 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.3 | 1.1 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.3 | 1.0 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.3 | 1.5 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.2 | 1.0 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.2 | 1.0 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.2 | 1.0 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.2 | 2.8 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.2 | 2.8 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.2 | 0.8 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.2 | 0.8 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.2 | 1.5 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.2 | 1.3 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.2 | 0.9 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.2 | 1.1 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.1 | 1.3 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 1.3 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.1 | 2.1 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 0.5 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 0.5 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.1 | 1.9 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 0.8 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.1 | 0.6 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.5 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.1 | 0.5 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.1 | 1.8 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.1 | 1.0 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 1.4 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 0.4 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 0.3 | GO:0016453 | C-acetyltransferase activity(GO:0016453) |
| 0.1 | 3.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 0.3 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 0.6 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 0.3 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 0.7 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.1 | 2.0 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.1 | 1.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 1.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 1.7 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 3.0 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 10.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 2.3 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 1.8 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.2 | GO:0000048 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.6 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 1.8 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.7 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.7 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.3 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.4 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 1.3 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 1.0 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 1.2 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 2.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.3 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.6 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 1.4 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.1 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 3.5 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 2.0 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 2.3 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 4.7 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.3 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 4.3 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 1.1 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.0 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 2.6 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 1.7 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 2.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 1.2 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.1 | 0.4 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 3.9 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 1.3 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.6 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 2.3 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.9 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.1 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 1.0 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.8 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.4 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.4 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.4 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.7 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.6 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.2 | 1.3 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 0.6 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 1.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 1.7 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.1 | 1.4 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 2.6 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 3.0 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 1.0 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 2.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.6 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 2.5 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 4.7 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 0.7 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.1 | 2.2 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 1.9 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 0.8 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 1.1 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.1 | 1.0 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 1.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.4 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.4 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.7 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.3 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.0 | 0.4 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 1.5 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.5 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 1.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 3.4 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 1.3 | REACTOME PI METABOLISM | Genes involved in PI Metabolism |
| 0.0 | 0.8 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.3 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.1 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.3 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 2.1 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.2 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |