Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
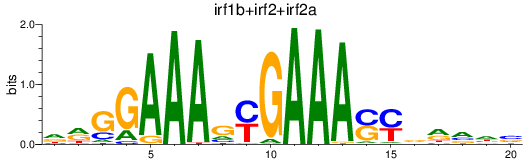
Results for irf1b+irf2+irf2a
Z-value: 1.68
Transcription factors associated with irf1b+irf2+irf2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
irf2a
|
ENSDARG00000007387 | interferon regulatory factor 2a |
|
irf2
|
ENSDARG00000040465 | interferon regulatory factor 2 |
|
irf1b
|
ENSDARG00000043249 | interferon regulatory factor 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| irf2a | dr11_v1_chr1_+_39865748_39865753 | 0.97 | 2.1e-05 | Click! |
| irf2 | dr11_v1_chr14_-_4121052_4121052 | -0.85 | 3.6e-03 | Click! |
| irf1b | dr11_v1_chr21_+_45626136_45626136 | -0.61 | 7.9e-02 | Click! |
Activity profile of irf1b+irf2+irf2a motif
Sorted Z-values of irf1b+irf2+irf2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_+_43668756 | 5.08 |
ENSDART00000112598
|
otud4
|
OTU deubiquitinase 4 |
| chr20_+_54295213 | 3.23 |
ENSDART00000074085
|
zp2.3
|
zona pellucida glycoprotein 2, tandem duplicate 3 |
| chr4_+_279669 | 2.70 |
ENSDART00000184884
|
CABZ01085275.1
|
|
| chr11_+_37638873 | 2.18 |
ENSDART00000186384
ENSDART00000184291 ENSDART00000131782 ENSDART00000140502 |
sh2d5
|
SH2 domain containing 5 |
| chr20_+_54309148 | 2.08 |
ENSDART00000099360
|
zp2.1
|
zona pellucida glycoprotein 2, tandem duplicate 1 |
| chr9_-_35633827 | 2.00 |
ENSDART00000077745
|
zp2l1
|
zona pellucida glycoprotein 2, like 1 |
| chr10_-_21542702 | 1.90 |
ENSDART00000146761
ENSDART00000134502 |
zgc:165539
|
zgc:165539 |
| chr18_-_38270430 | 1.89 |
ENSDART00000139519
|
caprin1b
|
cell cycle associated protein 1b |
| chr9_+_8401796 | 1.89 |
ENSDART00000180047
ENSDART00000132339 ENSDART00000137527 |
si:ch1073-75o15.3
|
si:ch1073-75o15.3 |
| chr14_-_45967712 | 1.88 |
ENSDART00000043751
ENSDART00000141357 |
macrod1
|
MACRO domain containing 1 |
| chr5_-_30382925 | 1.87 |
ENSDART00000125381
|
gig2o
|
grass carp reovirus (GCRV)-induced gene 2o |
| chr21_+_20901505 | 1.84 |
ENSDART00000132741
|
c7b
|
complement component 7b |
| chr9_-_20853439 | 1.83 |
ENSDART00000028247
ENSDART00000133321 |
gdap2
|
ganglioside induced differentiation associated protein 2 |
| chr13_-_6252498 | 1.80 |
ENSDART00000115157
|
tuba4l
|
tubulin, alpha 4 like |
| chr14_-_45967981 | 1.76 |
ENSDART00000188062
|
macrod1
|
MACRO domain containing 1 |
| chr18_-_12858016 | 1.74 |
ENSDART00000130343
|
parp12a
|
poly (ADP-ribose) polymerase family, member 12a |
| chr6_+_38896158 | 1.74 |
ENSDART00000029930
ENSDART00000131347 |
slc48a1b
|
solute carrier family 48 (heme transporter), member 1b |
| chr7_+_19600262 | 1.73 |
ENSDART00000007310
|
zgc:171731
|
zgc:171731 |
| chr3_-_3413669 | 1.72 |
ENSDART00000113517
ENSDART00000179861 ENSDART00000115331 |
zgc:171446
|
zgc:171446 |
| chr8_-_14604606 | 1.70 |
ENSDART00000090254
ENSDART00000188953 |
cep350
|
centrosomal protein 350 |
| chr10_+_44641599 | 1.68 |
ENSDART00000172128
|
sez6l
|
seizure related 6 homolog (mouse)-like |
| chr5_-_26247215 | 1.67 |
ENSDART00000136806
|
erap1b
|
endoplasmic reticulum aminopeptidase 1b |
| chr14_-_41467497 | 1.66 |
ENSDART00000181220
|
mid1ip1l
|
MID1 interacting protein 1, like |
| chr21_-_40557281 | 1.63 |
ENSDART00000172327
|
taok1b
|
TAO kinase 1b |
| chr14_+_36414856 | 1.59 |
ENSDART00000123343
ENSDART00000015761 |
neil3
|
nei-like DNA glycosylase 3 |
| chr15_-_18209672 | 1.57 |
ENSDART00000141508
ENSDART00000136280 |
btr16
|
bloodthirsty-related gene family, member 16 |
| chr13_-_37631092 | 1.57 |
ENSDART00000108855
|
si:dkey-188i13.7
|
si:dkey-188i13.7 |
| chr5_+_40835601 | 1.54 |
ENSDART00000147767
|
si:dkey-3h3.3
|
si:dkey-3h3.3 |
| chr3_-_36127234 | 1.53 |
ENSDART00000130917
|
coil
|
coilin p80 |
| chr5_-_57723929 | 1.49 |
ENSDART00000144237
|
gig2p
|
grass carp reovirus (GCRV)-induced gene 2p |
| chr11_+_1584747 | 1.45 |
ENSDART00000154583
|
si:dkey-40c23.2
|
si:dkey-40c23.2 |
| chr10_-_45229877 | 1.43 |
ENSDART00000169281
|
pargl
|
poly (ADP-ribose) glycohydrolase, like |
| chr7_-_54430505 | 1.42 |
ENSDART00000167905
|
ano1
|
anoctamin 1, calcium activated chloride channel |
| chr13_-_34858500 | 1.42 |
ENSDART00000184843
|
sptlc3
|
serine palmitoyltransferase, long chain base subunit 3 |
| chr18_+_14342326 | 1.41 |
ENSDART00000181013
ENSDART00000138372 |
si:dkey-246g23.2
|
si:dkey-246g23.2 |
| chr21_+_34992550 | 1.40 |
ENSDART00000109041
ENSDART00000135400 |
tmprss15
|
transmembrane protease, serine 15 |
| chr9_-_39547907 | 1.39 |
ENSDART00000163635
|
erbb4b
|
erb-b2 receptor tyrosine kinase 4b |
| chr2_-_42552666 | 1.39 |
ENSDART00000141399
|
dip2cb
|
disco-interacting protein 2 homolog Cb |
| chr18_-_38270077 | 1.38 |
ENSDART00000185546
|
caprin1b
|
cell cycle associated protein 1b |
| chr21_-_21514176 | 1.35 |
ENSDART00000031205
|
nectin3b
|
nectin cell adhesion molecule 3b |
| chr21_+_4256291 | 1.32 |
ENSDART00000148138
|
lrrc8aa
|
leucine rich repeat containing 8 VRAC subunit Aa |
| chr1_-_51266394 | 1.29 |
ENSDART00000164016
|
kif16ba
|
kinesin family member 16Ba |
| chr3_-_34586403 | 1.29 |
ENSDART00000151515
|
sept9a
|
septin 9a |
| chr4_-_26095755 | 1.28 |
ENSDART00000100611
ENSDART00000191266 |
si:ch211-244b2.3
|
si:ch211-244b2.3 |
| chr18_-_38270596 | 1.28 |
ENSDART00000098889
|
caprin1b
|
cell cycle associated protein 1b |
| chr15_+_25489406 | 1.27 |
ENSDART00000162482
|
zgc:152863
|
zgc:152863 |
| chr1_-_52201266 | 1.24 |
ENSDART00000143805
ENSDART00000023757 |
rab3da
|
RAB3D, member RAS oncogene family, a |
| chr18_+_13248956 | 1.24 |
ENSDART00000080709
|
plcg2
|
phospholipase C, gamma 2 |
| chr2_+_9757453 | 1.19 |
ENSDART00000168972
|
pcyt1aa
|
phosphate cytidylyltransferase 1, choline, alpha a |
| chr22_+_31023205 | 1.17 |
ENSDART00000111561
|
zmp:0000000735
|
zmp:0000000735 |
| chr17_-_35076730 | 1.15 |
ENSDART00000146590
|
mboat2a
|
membrane bound O-acyltransferase domain containing 2a |
| chr15_+_20352123 | 1.13 |
ENSDART00000011030
ENSDART00000163532 ENSDART00000169537 ENSDART00000161047 |
il15l
|
interleukin 15, like |
| chr12_+_17154655 | 1.12 |
ENSDART00000028003
|
ankrd22
|
ankyrin repeat domain 22 |
| chr7_+_55518519 | 1.12 |
ENSDART00000098476
ENSDART00000149915 |
cdt1
|
chromatin licensing and DNA replication factor 1 |
| chr13_-_42066299 | 1.12 |
ENSDART00000111536
|
rmdn2
|
regulator of microtubule dynamics 2 |
| chr9_-_14273652 | 1.10 |
ENSDART00000135458
|
abcb6b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6b |
| chr15_-_29162193 | 1.09 |
ENSDART00000138449
ENSDART00000099885 |
xaf1
|
XIAP associated factor 1 |
| chr1_-_55750208 | 1.08 |
ENSDART00000142244
|
dnajb1b
|
DnaJ (Hsp40) homolog, subfamily B, member 1b |
| chr22_-_8174244 | 1.07 |
ENSDART00000156571
|
si:ch73-44m9.3
|
si:ch73-44m9.3 |
| chr2_+_35603637 | 1.07 |
ENSDART00000147278
|
plk3
|
polo-like kinase 3 (Drosophila) |
| chr25_-_12803723 | 1.07 |
ENSDART00000158787
|
ca5a
|
carbonic anhydrase Va |
| chr7_+_40081630 | 1.06 |
ENSDART00000173559
|
zgc:112356
|
zgc:112356 |
| chr22_-_37565348 | 1.06 |
ENSDART00000149482
ENSDART00000104478 |
fxr1
|
fragile X mental retardation, autosomal homolog 1 |
| chr19_+_4066449 | 1.04 |
ENSDART00000162461
|
btr26
|
bloodthirsty-related gene family, member 26 |
| chr24_-_34680956 | 1.04 |
ENSDART00000171009
|
ctnna1
|
catenin (cadherin-associated protein), alpha 1 |
| chr8_-_19051906 | 1.04 |
ENSDART00000089024
|
sema6bb
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6Bb |
| chr11_+_1608348 | 1.02 |
ENSDART00000162438
|
si:dkey-40c23.3
|
si:dkey-40c23.3 |
| chr14_+_9485070 | 1.01 |
ENSDART00000161486
ENSDART00000137274 |
st3gal5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr14_+_12169979 | 1.01 |
ENSDART00000129953
|
rhogd
|
ras homolog gene family, member Gd |
| chr15_-_44077937 | 1.00 |
ENSDART00000110112
|
lamtor1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1 |
| chr21_+_31253048 | 0.99 |
ENSDART00000178521
ENSDART00000132317 ENSDART00000040190 |
asl
|
argininosuccinate lyase |
| chr20_+_49787584 | 0.99 |
ENSDART00000193458
ENSDART00000181511 ENSDART00000185850 ENSDART00000185613 ENSDART00000191671 |
CABZ01078261.1
|
|
| chr18_+_19990412 | 0.99 |
ENSDART00000155054
ENSDART00000090310 |
pias1b
|
protein inhibitor of activated STAT, 1b |
| chr6_-_1566407 | 0.98 |
ENSDART00000112118
|
trim107
|
tripartite motif containing 107 |
| chr22_-_5958066 | 0.97 |
ENSDART00000145821
|
si:rp71-36a1.3
|
si:rp71-36a1.3 |
| chr8_-_16725959 | 0.96 |
ENSDART00000183593
|
depdc1a
|
DEP domain containing 1a |
| chr23_+_45229198 | 0.96 |
ENSDART00000172445
|
ttc39b
|
tetratricopeptide repeat domain 39B |
| chr2_-_32688905 | 0.95 |
ENSDART00000041146
|
nrbp2a
|
nuclear receptor binding protein 2a |
| chr5_+_57726425 | 0.95 |
ENSDART00000134684
|
fdxacb1
|
ferredoxin-fold anticodon binding domain containing 1 |
| chr19_-_35492693 | 0.94 |
ENSDART00000135838
ENSDART00000051745 ENSDART00000177052 |
ptp4a2b
|
protein tyrosine phosphatase type IVA, member 2b |
| chr13_+_7387822 | 0.93 |
ENSDART00000148240
|
exoc3l4
|
exocyst complex component 3-like 4 |
| chr21_+_40695345 | 0.93 |
ENSDART00000143594
|
ccdc82
|
coiled-coil domain containing 82 |
| chr10_+_5954787 | 0.93 |
ENSDART00000161887
ENSDART00000160345 ENSDART00000190046 |
map3k1
|
mitogen-activated protein kinase kinase kinase 1, E3 ubiquitin protein ligase |
| chr18_-_6534357 | 0.93 |
ENSDART00000192886
|
ddx11
|
DEAD/H (Asp-Glu-Ala-Asp/His) box helicase 11 |
| chr15_-_25126842 | 0.93 |
ENSDART00000193670
|
vps53
|
vacuolar protein sorting 53 homolog (S. cerevisiae) |
| chr3_+_36127287 | 0.92 |
ENSDART00000058605
ENSDART00000182500 |
scpep1
|
serine carboxypeptidase 1 |
| chr8_-_51367298 | 0.91 |
ENSDART00000060628
|
chmp7
|
charged multivesicular body protein 7 |
| chr13_+_7578111 | 0.91 |
ENSDART00000175431
|
gbf1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
| chr23_-_21758253 | 0.91 |
ENSDART00000046613
|
vps13d
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr15_-_1484795 | 0.91 |
ENSDART00000129356
|
si:dkeyp-97b10.3
|
si:dkeyp-97b10.3 |
| chr5_-_37116265 | 0.90 |
ENSDART00000057613
|
il13ra2
|
interleukin 13 receptor, alpha 2 |
| chr17_-_42799104 | 0.90 |
ENSDART00000154755
|
prkd3
|
protein kinase D3 |
| chr3_-_40136743 | 0.90 |
ENSDART00000149546
|
myo15aa
|
myosin XVAa |
| chr6_-_1566581 | 0.90 |
ENSDART00000192993
|
trim107
|
tripartite motif containing 107 |
| chr25_-_12804450 | 0.89 |
ENSDART00000169717
|
ca5a
|
carbonic anhydrase Va |
| chr18_-_6534516 | 0.89 |
ENSDART00000009217
|
ddx11
|
DEAD/H (Asp-Glu-Ala-Asp/His) box helicase 11 |
| chr9_-_40683722 | 0.88 |
ENSDART00000141979
ENSDART00000181228 |
bard1
|
BRCA1 associated RING domain 1 |
| chr2_-_40889465 | 0.88 |
ENSDART00000192631
ENSDART00000180824 |
uggt1
|
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr11_-_30341431 | 0.88 |
ENSDART00000078378
|
cflara
|
CASP8 and FADD-like apoptosis regulator a |
| chr1_-_55058795 | 0.88 |
ENSDART00000187293
|
peli1a
|
pellino E3 ubiquitin protein ligase 1a |
| chr16_+_53710416 | 0.87 |
ENSDART00000155929
|
nod1
|
nucleotide-binding oligomerization domain containing 1 |
| chr15_+_14592624 | 0.87 |
ENSDART00000162350
|
FBXO46
|
si:dkey-114g7.4 |
| chr11_-_11791718 | 0.87 |
ENSDART00000180476
|
cdc6
|
cell division cycle 6 homolog (S. cerevisiae) |
| chr16_+_26439939 | 0.87 |
ENSDART00000143073
|
trim35-28
|
tripartite motif containing 35-28 |
| chr24_+_39027481 | 0.86 |
ENSDART00000085565
|
capn15
|
calpain 15 |
| chr15_-_14083028 | 0.86 |
ENSDART00000147796
ENSDART00000043492 ENSDART00000133080 |
trappc6bl
|
trafficking protein particle complex 6b-like |
| chr17_+_25871304 | 0.85 |
ENSDART00000185143
|
wapla
|
WAPL cohesin release factor a |
| chr17_+_50798484 | 0.85 |
ENSDART00000125787
|
stxbp6
|
syntaxin binding protein 6 (amisyn) |
| chr19_+_15485287 | 0.85 |
ENSDART00000182797
|
pdik1l
|
PDLIM1 interacting kinase 1 like |
| chr15_-_15227541 | 0.85 |
ENSDART00000184787
|
rrp8
|
ribosomal RNA processing 8, methyltransferase, homolog (yeast) |
| chr15_+_17100697 | 0.85 |
ENSDART00000183565
ENSDART00000123197 |
relb
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr8_-_49499457 | 0.84 |
ENSDART00000098326
|
opn7d
|
opsin 7, group member d |
| chr17_-_7218481 | 0.84 |
ENSDART00000181967
|
SAMD5
|
sterile alpha motif domain containing 5 |
| chr1_-_12397258 | 0.84 |
ENSDART00000144596
|
sclt1
|
sodium channel and clathrin linker 1 |
| chr22_-_6941098 | 0.84 |
ENSDART00000105864
|
zgc:171500
|
zgc:171500 |
| chr15_-_16384184 | 0.83 |
ENSDART00000154504
|
fam222bb
|
family with sequence similarity 222, member Bb |
| chr5_+_36768674 | 0.83 |
ENSDART00000146854
|
mark4a
|
MAP/microtubule affinity-regulating kinase 4a |
| chr14_-_33328031 | 0.83 |
ENSDART00000137997
|
sept6
|
septin 6 |
| chr20_+_44582318 | 0.82 |
ENSDART00000149000
ENSDART00000149775 ENSDART00000085416 |
atad2b
|
ATPase family, AAA domain containing 2B |
| chr19_+_4856351 | 0.81 |
ENSDART00000093402
|
cdk12
|
cyclin-dependent kinase 12 |
| chr11_-_20096018 | 0.81 |
ENSDART00000030420
|
ogfrl2
|
opioid growth factor receptor-like 2 |
| chr6_-_57539141 | 0.81 |
ENSDART00000156967
|
itcha
|
itchy E3 ubiquitin protein ligase a |
| chr25_-_27665978 | 0.81 |
ENSDART00000123590
|
si:ch211-91p5.3
|
si:ch211-91p5.3 |
| chr20_-_26588736 | 0.80 |
ENSDART00000134337
|
exoc2
|
exocyst complex component 2 |
| chr21_-_43428040 | 0.80 |
ENSDART00000148325
|
stk26
|
serine/threonine protein kinase 26 |
| chr22_+_1440702 | 0.80 |
ENSDART00000165677
|
si:dkeyp-53d3.3
|
si:dkeyp-53d3.3 |
| chr6_+_9793495 | 0.80 |
ENSDART00000108524
|
als2b
|
amyotrophic lateral sclerosis 2b (juvenile) |
| chr19_+_7115223 | 0.80 |
ENSDART00000001359
|
psmb12
|
proteasome subunit beta 12 |
| chr4_+_59061652 | 0.80 |
ENSDART00000150447
|
znf1127
|
zinc finger protein 1127 |
| chr6_+_13045885 | 0.80 |
ENSDART00000104757
|
casp8
|
caspase 8, apoptosis-related cysteine peptidase |
| chr6_+_30430591 | 0.80 |
ENSDART00000108943
|
shroom2a
|
shroom family member 2a |
| chr8_-_11067079 | 0.79 |
ENSDART00000181986
|
dennd2c
|
DENN/MADD domain containing 2C |
| chr16_-_40459104 | 0.79 |
ENSDART00000032389
|
plekhf2
|
pleckstrin homology domain containing, family F (with FYVE domain) member 2 |
| chr20_-_23254876 | 0.79 |
ENSDART00000141510
|
ociad1
|
OCIA domain containing 1 |
| chr19_+_20178978 | 0.79 |
ENSDART00000145115
ENSDART00000151175 |
tra2a
|
transformer 2 alpha homolog |
| chr3_+_51684963 | 0.79 |
ENSDART00000091180
ENSDART00000183711 ENSDART00000159493 |
baiap2a
|
BAI1-associated protein 2a |
| chr15_+_44228917 | 0.78 |
ENSDART00000159630
|
CU929391.3
|
|
| chr1_+_39865748 | 0.77 |
ENSDART00000131954
ENSDART00000130270 |
irf2a
|
interferon regulatory factor 2a |
| chr4_+_20486041 | 0.77 |
ENSDART00000017572
|
ints13
|
integrator complex subunit 13 |
| chr5_+_22406672 | 0.76 |
ENSDART00000141385
|
si:dkey-27p18.3
|
si:dkey-27p18.3 |
| chr22_-_7461603 | 0.76 |
ENSDART00000170630
|
BX511034.6
|
|
| chr14_-_25078569 | 0.75 |
ENSDART00000172802
ENSDART00000173345 ENSDART00000135004 |
matr3l1.1
|
matrin 3-like 1.1 |
| chr7_+_44593756 | 0.75 |
ENSDART00000125365
|
si:ch211-189a15.5
|
si:ch211-189a15.5 |
| chr1_-_45584407 | 0.74 |
ENSDART00000149155
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr2_+_42005475 | 0.74 |
ENSDART00000056461
|
gbp2
|
guanylate binding protein 2 |
| chr11_+_2596667 | 0.73 |
ENSDART00000175330
|
dnajc14
|
DnaJ (Hsp40) homolog, subfamily C, member 14 |
| chr18_+_14684115 | 0.73 |
ENSDART00000108469
|
spata2l
|
spermatogenesis associated 2-like |
| chr23_+_2825940 | 0.73 |
ENSDART00000135781
|
plcg1
|
phospholipase C, gamma 1 |
| chr5_+_17780475 | 0.72 |
ENSDART00000110783
ENSDART00000115227 |
chfr
|
checkpoint with forkhead and ring finger domains, E3 ubiquitin protein ligase |
| chr10_+_35152928 | 0.72 |
ENSDART00000063418
|
nsun5
|
NOP2/Sun domain family, member 5 |
| chr16_+_26439518 | 0.71 |
ENSDART00000041787
|
trim35-28
|
tripartite motif containing 35-28 |
| chr1_-_524433 | 0.71 |
ENSDART00000147610
|
si:ch73-41e3.7
|
si:ch73-41e3.7 |
| chr21_+_19319804 | 0.71 |
ENSDART00000063621
|
abraxas2a
|
abraxas 2a, BRISC complex subunit |
| chr13_+_11550454 | 0.70 |
ENSDART00000034935
ENSDART00000166908 |
desi2
|
desumoylating isopeptidase 2 |
| chr8_-_14080534 | 0.70 |
ENSDART00000042867
|
dedd
|
death effector domain containing |
| chr7_-_40656148 | 0.70 |
ENSDART00000142315
|
nom1
|
nucleolar protein with MIF4G domain 1 |
| chr6_-_1566186 | 0.69 |
ENSDART00000156305
|
trim107
|
tripartite motif containing 107 |
| chr16_-_50952266 | 0.69 |
ENSDART00000165408
|
si:dkeyp-97a10.3
|
si:dkeyp-97a10.3 |
| chr5_+_45895791 | 0.68 |
ENSDART00000141039
|
polk
|
polymerase (DNA directed) kappa |
| chr6_-_42949184 | 0.67 |
ENSDART00000147208
|
edem1
|
ER degradation enhancer, mannosidase alpha-like 1 |
| chr13_-_31938512 | 0.67 |
ENSDART00000026726
ENSDART00000182666 |
diexf
|
digestive organ expansion factor homolog |
| chr15_-_36249087 | 0.67 |
ENSDART00000157190
|
ostn
|
osteocrin |
| chr11_-_45385803 | 0.67 |
ENSDART00000173329
|
trappc10
|
trafficking protein particle complex 10 |
| chr23_-_36449111 | 0.66 |
ENSDART00000110478
|
zgc:174906
|
zgc:174906 |
| chr14_+_31618982 | 0.66 |
ENSDART00000026195
|
slc9a6a
|
solute carrier family 9, subfamily A (NHE6, cation proton antiporter 6), member 6a |
| chr10_+_7718156 | 0.65 |
ENSDART00000189101
|
ggcx
|
gamma-glutamyl carboxylase |
| chr22_+_1786230 | 0.65 |
ENSDART00000169318
ENSDART00000164948 |
znf1154
|
zinc finger protein 1154 |
| chr14_-_25095808 | 0.65 |
ENSDART00000184244
|
matr3l1.1
|
matrin 3-like 1.1 |
| chr22_-_8006342 | 0.65 |
ENSDART00000162028
|
sc:d217
|
sc:d217 |
| chr18_-_44935174 | 0.64 |
ENSDART00000081025
|
pex16
|
peroxisomal biogenesis factor 16 |
| chr4_-_9191220 | 0.64 |
ENSDART00000156919
|
hcfc2
|
host cell factor C2 |
| chr19_+_9111550 | 0.64 |
ENSDART00000088336
|
setdb1a
|
SET domain, bifurcated 1a |
| chr21_+_25120546 | 0.63 |
ENSDART00000149507
|
ddx10
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 10 |
| chr22_+_2512154 | 0.63 |
ENSDART00000097363
|
zgc:173726
|
zgc:173726 |
| chr21_-_1644414 | 0.63 |
ENSDART00000105736
ENSDART00000124904 |
zgc:152948
|
zgc:152948 |
| chr22_+_29990448 | 0.63 |
ENSDART00000165313
|
si:dkey-286j15.3
|
si:dkey-286j15.3 |
| chr20_-_4793450 | 0.63 |
ENSDART00000053870
|
galca
|
galactosylceramidase a |
| chr20_+_32481348 | 0.63 |
ENSDART00000185018
|
ostm1
|
osteopetrosis associated transmembrane protein 1 |
| chr5_+_52625975 | 0.62 |
ENSDART00000170341
ENSDART00000168317 |
apba1a
|
amyloid beta (A4) precursor protein-binding, family A, member 1a |
| chr7_-_19613388 | 0.62 |
ENSDART00000173664
|
si:ch211-212k18.13
|
si:ch211-212k18.13 |
| chr6_+_29861288 | 0.62 |
ENSDART00000166782
|
dlg1
|
discs, large homolog 1 (Drosophila) |
| chr19_+_38168006 | 0.62 |
ENSDART00000087662
ENSDART00000177759 |
phf14
|
PHD finger protein 14 |
| chr24_-_10828560 | 0.62 |
ENSDART00000132282
|
fam49bb
|
family with sequence similarity 49, member Bb |
| chr5_+_38612134 | 0.62 |
ENSDART00000135600
ENSDART00000181144 |
si:ch211-271e10.2
|
si:ch211-271e10.2 |
| chr25_-_19574146 | 0.62 |
ENSDART00000156811
|
si:ch211-59o9.10
|
si:ch211-59o9.10 |
| chr23_-_36884012 | 0.62 |
ENSDART00000137282
|
ube2j2
|
ubiquitin-conjugating enzyme E2, J2 (UBC6 homolog, yeast) |
| chr16_+_30117798 | 0.61 |
ENSDART00000135723
ENSDART00000000198 |
sema6e
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6E |
| chr24_+_12945803 | 0.61 |
ENSDART00000005105
|
psme1
|
proteasome activator subunit 1 |
| chr20_-_38525467 | 0.61 |
ENSDART00000061417
|
si:ch211-245h14.1
|
si:ch211-245h14.1 |
| chr13_-_33321058 | 0.61 |
ENSDART00000112084
|
si:dkey-71p21.13
|
si:dkey-71p21.13 |
| chr6_+_9793791 | 0.60 |
ENSDART00000149896
|
als2b
|
amyotrophic lateral sclerosis 2b (juvenile) |
| chr11_-_19775182 | 0.60 |
ENSDART00000037894
|
namptb
|
nicotinamide phosphoribosyltransferase b |
| chr19_-_5865766 | 0.60 |
ENSDART00000191007
|
LO018585.1
|
|
| chr15_-_15965456 | 0.59 |
ENSDART00000154579
|
synrg
|
synergin, gamma |
| chr17_-_15149192 | 0.59 |
ENSDART00000180511
ENSDART00000103405 |
gch1
|
GTP cyclohydrolase 1 |
| chr13_-_33227411 | 0.59 |
ENSDART00000057386
|
golga5
|
golgin A5 |
| chr5_-_4923473 | 0.59 |
ENSDART00000134585
|
zbtb43
|
zinc finger and BTB domain containing 43 |
| chr7_+_39416336 | 0.58 |
ENSDART00000171783
|
CT030188.1
|
|
| chr9_+_22375331 | 0.58 |
ENSDART00000090907
|
dgkg
|
diacylglycerol kinase, gamma |
| chr15_-_17169935 | 0.58 |
ENSDART00000110111
|
cul5a
|
cullin 5a |
Network of associatons between targets according to the STRING database.
First level regulatory network of irf1b+irf2+irf2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.1 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.6 | 1.8 | GO:1901836 | regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901836) |
| 0.4 | 1.6 | GO:0048308 | organelle inheritance(GO:0048308) Golgi inheritance(GO:0048313) |
| 0.3 | 1.5 | GO:0030576 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.3 | 0.9 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.3 | 2.8 | GO:0015886 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.3 | 0.8 | GO:0043903 | regulation of symbiosis, encompassing mutualism through parasitism(GO:0043903) |
| 0.2 | 1.0 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.2 | 0.7 | GO:0035477 | regulation of angioblast cell migration involved in selective angioblast sprouting(GO:0035477) |
| 0.2 | 2.3 | GO:0019883 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.2 | 1.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.2 | 1.2 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.2 | 0.9 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.2 | 1.0 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.2 | 1.7 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.2 | 0.8 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.2 | 0.9 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.1 | 0.6 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.1 | 0.9 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.1 | 1.2 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.1 | 2.0 | GO:0000076 | DNA replication checkpoint(GO:0000076) |
| 0.1 | 0.5 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.1 | 0.5 | GO:0071788 | endoplasmic reticulum tubular network maintenance(GO:0071788) |
| 0.1 | 1.4 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.1 | 0.6 | GO:0014005 | microglia development(GO:0014005) |
| 0.1 | 0.7 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.1 | 1.4 | GO:0090309 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.1 | 0.7 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.1 | 0.9 | GO:0035188 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.1 | 0.6 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 0.6 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 0.4 | GO:0070587 | regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 0.1 | 0.8 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 0.3 | GO:1903292 | protein localization to Golgi membrane(GO:1903292) |
| 0.1 | 0.4 | GO:1903573 | negative regulation of response to endoplasmic reticulum stress(GO:1903573) |
| 0.1 | 0.5 | GO:0045337 | farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.1 | 1.2 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.1 | 0.9 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.1 | 1.1 | GO:0036230 | granulocyte activation(GO:0036230) neutrophil activation(GO:0042119) |
| 0.1 | 1.7 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 0.9 | GO:1902042 | regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902041) negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.1 | 0.2 | GO:0051793 | medium-chain fatty acid metabolic process(GO:0051791) medium-chain fatty acid catabolic process(GO:0051793) |
| 0.1 | 0.6 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.1 | 0.2 | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation(GO:1901533) |
| 0.1 | 0.7 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.1 | 0.7 | GO:0034627 | 'de novo' NAD biosynthetic process from tryptophan(GO:0034354) 'de novo' NAD biosynthetic process(GO:0034627) |
| 0.1 | 0.3 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.1 | 0.7 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 | 0.4 | GO:0002504 | antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.1 | 0.3 | GO:0044819 | mitotic G1 DNA damage checkpoint(GO:0031571) mitotic G1/S transition checkpoint(GO:0044819) |
| 0.1 | 1.8 | GO:0019835 | cytolysis(GO:0019835) |
| 0.1 | 0.2 | GO:0071514 | genetic imprinting(GO:0071514) |
| 0.1 | 0.3 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.1 | 0.4 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 0.8 | GO:0045176 | asymmetric protein localization(GO:0008105) apical protein localization(GO:0045176) |
| 0.1 | 0.8 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.7 | GO:0097369 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 0.9 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 | 0.8 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 | 0.3 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.1 | 0.9 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.1 | 1.4 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.1 | 1.7 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.1 | 0.2 | GO:0019884 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) antigen processing and presentation of exogenous antigen(GO:0019884) antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.0 | 0.8 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.2 | GO:0071267 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.0 | 1.1 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 2.8 | GO:0071166 | ribonucleoprotein complex localization(GO:0071166) |
| 0.0 | 1.4 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.5 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 2.4 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.3 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.8 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.2 | GO:0043703 | retinal cone cell fate determination(GO:0042671) eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate determination(GO:0043703) retinal cone cell fate commitment(GO:0046551) photoreceptor cell fate commitment(GO:0046552) camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.0 | 0.8 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.0 | 0.3 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.0 | 0.4 | GO:0035479 | angioblast cell migration from lateral mesoderm to midline(GO:0035479) |
| 0.0 | 0.6 | GO:1902807 | negative regulation of cell cycle G1/S phase transition(GO:1902807) negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.3 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.1 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter in response to calcium ion(GO:0061400) positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 1.4 | GO:0009225 | nucleotide-sugar metabolic process(GO:0009225) |
| 0.0 | 0.4 | GO:0031269 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 1.6 | GO:0006289 | nucleotide-excision repair(GO:0006289) |
| 0.0 | 2.0 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.3 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.4 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 1.0 | GO:0071910 | determination of liver left/right asymmetry(GO:0071910) |
| 0.0 | 0.2 | GO:0086013 | membrane repolarization during cardiac muscle cell action potential(GO:0086013) |
| 0.0 | 0.8 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 3.6 | GO:0017148 | negative regulation of translation(GO:0017148) |
| 0.0 | 1.0 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.2 | GO:0048662 | regulation of smooth muscle cell proliferation(GO:0048660) negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.0 | 0.8 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 1.5 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 1.2 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.5 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 1.2 | GO:0009395 | phospholipid catabolic process(GO:0009395) |
| 0.0 | 0.3 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.6 | GO:2000045 | regulation of G1/S transition of mitotic cell cycle(GO:2000045) |
| 0.0 | 0.2 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.3 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.6 | GO:0034112 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.0 | 0.9 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.5 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 0.5 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.4 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.7 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.3 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.0 | 0.3 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.1 | GO:0031179 | peptide modification(GO:0031179) |
| 0.0 | 0.2 | GO:1904356 | regulation of telomere maintenance via telomere lengthening(GO:1904356) |
| 0.0 | 0.1 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.0 | 0.3 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.5 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.0 | 3.1 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 0.0 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.0 | 0.5 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.4 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 1.9 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 9.7 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 2.3 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.9 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 1.2 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.4 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 0.6 | GO:0007031 | peroxisome organization(GO:0007031) |
| 0.0 | 0.2 | GO:0070307 | lens fiber cell development(GO:0070307) lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.2 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.0 | 0.6 | GO:0006626 | protein targeting to mitochondrion(GO:0006626) |
| 0.0 | 0.1 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 0.0 | 0.4 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 1.3 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.4 | GO:0051084 | 'de novo' protein folding(GO:0006458) 'de novo' posttranslational protein folding(GO:0051084) chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.4 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 0.2 | GO:0060034 | notochord cell differentiation(GO:0060034) |
| 0.0 | 1.4 | GO:0000819 | sister chromatid segregation(GO:0000819) |
| 0.0 | 0.7 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.0 | 1.0 | GO:0001894 | tissue homeostasis(GO:0001894) |
| 0.0 | 0.3 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.9 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.0 | 0.7 | GO:0042274 | ribosomal small subunit biogenesis(GO:0042274) |
| 0.0 | 0.2 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.0 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.0 | 0.2 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.4 | GO:0051028 | mRNA transport(GO:0051028) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0044326 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.2 | 1.2 | GO:1990071 | TRAPPII protein complex(GO:1990071) |
| 0.2 | 0.6 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.2 | 0.9 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.2 | 1.0 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.2 | 0.9 | GO:0000938 | GARP complex(GO:0000938) |
| 0.2 | 0.6 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 0.9 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.1 | 0.7 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.1 | 1.4 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 0.6 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 0.9 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 0.5 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.1 | 1.8 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.9 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.1 | 1.1 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 0.8 | GO:0019908 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.1 | 0.4 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.1 | 1.5 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 0.7 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.3 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 2.0 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 0.9 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 1.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 0.9 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 1.2 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 0.4 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.1 | 2.1 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 1.0 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.3 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 0.5 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 1.8 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.9 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.3 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.4 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.3 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.6 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.6 | GO:0030130 | trans-Golgi network transport vesicle membrane(GO:0012510) clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.2 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.5 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.6 | GO:0000784 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 0.2 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 1.3 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.6 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.3 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.0 | 3.0 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 1.3 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.4 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.6 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.6 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.6 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.8 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.2 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.1 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 1.4 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 1.3 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 1.5 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.6 | GO:0055037 | recycling endosome(GO:0055037) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.4 | GO:0004649 | poly(ADP-ribose) glycohydrolase activity(GO:0004649) |
| 0.4 | 2.8 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.3 | 1.0 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.3 | 5.1 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.3 | 1.0 | GO:0016436 | rRNA (uridine) methyltransferase activity(GO:0016436) |
| 0.3 | 1.5 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.3 | 1.1 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.2 | 1.0 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.2 | 1.2 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.2 | 1.4 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.2 | 0.9 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.2 | 0.9 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.2 | 0.5 | GO:0046978 | TAP1 binding(GO:0046978) |
| 0.2 | 0.5 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.2 | 0.7 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.2 | 0.8 | GO:0051748 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.2 | 0.5 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 0.9 | GO:1900750 | oligopeptide binding(GO:1900750) |
| 0.1 | 0.7 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.1 | 0.4 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 0.6 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.1 | 1.3 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 0.4 | GO:0051500 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.1 | 0.4 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.1 | 1.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 0.3 | GO:0015462 | protein-transmembrane transporting ATPase activity(GO:0015462) |
| 0.1 | 0.3 | GO:0047760 | medium-chain fatty acid-CoA ligase activity(GO:0031956) butyrate-CoA ligase activity(GO:0047760) |
| 0.1 | 1.6 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.1 | 1.6 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.1 | 0.2 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) peptide-transporting ATPase activity(GO:0015440) TAP binding(GO:0046977) |
| 0.1 | 0.6 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.6 | GO:0005549 | olfactory receptor activity(GO:0004984) odorant binding(GO:0005549) |
| 0.1 | 4.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 0.3 | GO:1902945 | metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902945) |
| 0.1 | 0.5 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 1.2 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.1 | 1.0 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 0.3 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 0.6 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 2.0 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.3 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.1 | 0.5 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 0.7 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.1 | 0.3 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.1 | 0.5 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.1 | 0.4 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 1.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 2.0 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.1 | 1.2 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 0.8 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 1.7 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.6 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.4 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 0.3 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.3 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 1.9 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.3 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.4 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.4 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 1.3 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.8 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.9 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.8 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.3 | GO:0050664 | oxidoreductase activity, acting on NAD(P)H, oxygen as acceptor(GO:0050664) |
| 0.0 | 0.5 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.5 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.2 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 1.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.5 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 1.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 4.0 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 1.2 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 1.1 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.0 | 0.2 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.9 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 1.5 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 1.0 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.7 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0036374 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 1.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.6 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.0 | 0.5 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.4 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 7.3 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.4 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.4 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 1.8 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.2 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 2.0 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 1.1 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.6 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.4 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 1.8 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 2.0 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 0.2 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.7 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 0.9 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 0.6 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 1.8 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 1.2 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.9 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.7 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 1.3 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 1.3 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 1.2 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 1.8 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.4 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.8 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.6 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.3 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.6 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.2 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.2 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.2 | 2.0 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 2.0 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.1 | 1.6 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 0.9 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 0.6 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 0.7 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 0.5 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 2.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 0.7 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 1.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.6 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.5 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.6 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 1.2 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 1.1 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.8 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.9 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.9 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.5 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.0 | 0.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.8 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 1.2 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.5 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.7 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.4 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.4 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.7 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.2 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.2 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.0 | 0.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.3 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.9 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.3 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |