Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
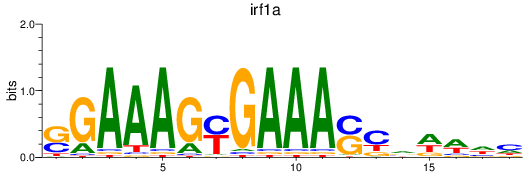
Results for irf1a
Z-value: 0.74
Transcription factors associated with irf1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
irf1a
|
ENSDARG00000043492 | interferon regulatory factor 1a |
Activity profile of irf1a motif
Sorted Z-values of irf1a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_279669 | 1.54 |
ENSDART00000184884
|
CABZ01085275.1
|
|
| chr3_-_3398383 | 1.45 |
ENSDART00000047865
|
si:dkey-46g23.2
|
si:dkey-46g23.2 |
| chr11_+_37638873 | 1.44 |
ENSDART00000186384
ENSDART00000184291 ENSDART00000131782 ENSDART00000140502 |
sh2d5
|
SH2 domain containing 5 |
| chr5_-_26247215 | 1.27 |
ENSDART00000136806
|
erap1b
|
endoplasmic reticulum aminopeptidase 1b |
| chr15_-_29162193 | 1.27 |
ENSDART00000138449
ENSDART00000099885 |
xaf1
|
XIAP associated factor 1 |
| chr18_+_14329231 | 1.26 |
ENSDART00000151641
|
zgc:173742
|
zgc:173742 |
| chr18_-_12858016 | 1.23 |
ENSDART00000130343
|
parp12a
|
poly (ADP-ribose) polymerase family, member 12a |
| chr2_-_38287987 | 1.10 |
ENSDART00000185329
ENSDART00000061677 |
si:ch211-14a17.6
|
si:ch211-14a17.6 |
| chr5_-_57723929 | 1.06 |
ENSDART00000144237
|
gig2p
|
grass carp reovirus (GCRV)-induced gene 2p |
| chr7_+_19600262 | 1.04 |
ENSDART00000007310
|
zgc:171731
|
zgc:171731 |
| chr23_+_43668756 | 1.00 |
ENSDART00000112598
|
otud4
|
OTU deubiquitinase 4 |
| chr6_+_38896158 | 0.95 |
ENSDART00000029930
ENSDART00000131347 |
slc48a1b
|
solute carrier family 48 (heme transporter), member 1b |
| chr3_-_34586403 | 0.95 |
ENSDART00000151515
|
sept9a
|
septin 9a |
| chr14_+_36414856 | 0.91 |
ENSDART00000123343
ENSDART00000015761 |
neil3
|
nei-like DNA glycosylase 3 |
| chr7_-_54430505 | 0.89 |
ENSDART00000167905
|
ano1
|
anoctamin 1, calcium activated chloride channel |
| chr10_-_1961576 | 0.86 |
ENSDART00000042441
|
tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr10_-_45229877 | 0.86 |
ENSDART00000169281
|
pargl
|
poly (ADP-ribose) glycohydrolase, like |
| chr18_-_38270430 | 0.83 |
ENSDART00000139519
|
caprin1b
|
cell cycle associated protein 1b |
| chr5_-_26247671 | 0.81 |
ENSDART00000145187
|
erap1b
|
endoplasmic reticulum aminopeptidase 1b |
| chr5_-_26247973 | 0.81 |
ENSDART00000098527
|
erap1b
|
endoplasmic reticulum aminopeptidase 1b |
| chr9_-_14273652 | 0.78 |
ENSDART00000135458
|
abcb6b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6b |
| chr5_-_38248347 | 0.76 |
ENSDART00000084917
ENSDART00000139479 |
slc12a9
|
solute carrier family 12, member 9 |
| chr12_-_13549538 | 0.75 |
ENSDART00000133895
|
ghdc
|
GH3 domain containing |
| chr1_-_52201266 | 0.72 |
ENSDART00000143805
ENSDART00000023757 |
rab3da
|
RAB3D, member RAS oncogene family, a |
| chr5_-_31856681 | 0.70 |
ENSDART00000187817
|
pkn3
|
protein kinase N3 |
| chr3_-_36127234 | 0.70 |
ENSDART00000130917
|
coil
|
coilin p80 |
| chr9_-_20853439 | 0.69 |
ENSDART00000028247
ENSDART00000133321 |
gdap2
|
ganglioside induced differentiation associated protein 2 |
| chr18_+_14684115 | 0.68 |
ENSDART00000108469
|
spata2l
|
spermatogenesis associated 2-like |
| chr7_-_60351876 | 0.68 |
ENSDART00000098563
|
plcb3
|
phospholipase C, beta 3 (phosphatidylinositol-specific) |
| chr23_+_20705849 | 0.65 |
ENSDART00000079538
|
ccdc30
|
coiled-coil domain containing 30 |
| chr2_-_42552666 | 0.64 |
ENSDART00000141399
|
dip2cb
|
disco-interacting protein 2 homolog Cb |
| chr1_-_52790724 | 0.63 |
ENSDART00000139577
ENSDART00000100937 |
patl1
|
protein associated with topoisomerase II homolog 1 (yeast) |
| chr8_-_19051906 | 0.63 |
ENSDART00000089024
|
sema6bb
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6Bb |
| chr9_+_22631672 | 0.63 |
ENSDART00000101770
ENSDART00000126015 ENSDART00000145005 |
etv5a
|
ets variant 5a |
| chr15_+_20352123 | 0.62 |
ENSDART00000011030
ENSDART00000163532 ENSDART00000169537 ENSDART00000161047 |
il15l
|
interleukin 15, like |
| chr4_+_59061652 | 0.62 |
ENSDART00000150447
|
znf1127
|
zinc finger protein 1127 |
| chr17_+_25833947 | 0.62 |
ENSDART00000044328
ENSDART00000154604 |
acss1
|
acyl-CoA synthetase short chain family member 1 |
| chr16_-_40459104 | 0.62 |
ENSDART00000032389
|
plekhf2
|
pleckstrin homology domain containing, family F (with FYVE domain) member 2 |
| chr1_-_55058795 | 0.61 |
ENSDART00000187293
|
peli1a
|
pellino E3 ubiquitin protein ligase 1a |
| chr18_-_6534357 | 0.60 |
ENSDART00000192886
|
ddx11
|
DEAD/H (Asp-Glu-Ala-Asp/His) box helicase 11 |
| chr8_-_5847533 | 0.59 |
ENSDART00000192489
|
CABZ01102147.1
|
|
| chr19_+_4066449 | 0.59 |
ENSDART00000162461
|
btr26
|
bloodthirsty-related gene family, member 26 |
| chr6_+_29861288 | 0.59 |
ENSDART00000166782
|
dlg1
|
discs, large homolog 1 (Drosophila) |
| chr16_+_26439939 | 0.57 |
ENSDART00000143073
|
trim35-28
|
tripartite motif containing 35-28 |
| chr19_+_14115838 | 0.57 |
ENSDART00000192057
|
kdf1b
|
keratinocyte differentiation factor 1b |
| chr7_-_19369002 | 0.57 |
ENSDART00000165680
|
ntn4
|
netrin 4 |
| chr17_+_18810904 | 0.57 |
ENSDART00000130899
|
si:dkey-288a3.2
|
si:dkey-288a3.2 |
| chr5_-_31857345 | 0.56 |
ENSDART00000112546
|
pkn3
|
protein kinase N3 |
| chr20_-_31497300 | 0.56 |
ENSDART00000046841
|
sash1a
|
SAM and SH3 domain containing 1a |
| chr18_-_38270077 | 0.56 |
ENSDART00000185546
|
caprin1b
|
cell cycle associated protein 1b |
| chr18_-_6534516 | 0.55 |
ENSDART00000009217
|
ddx11
|
DEAD/H (Asp-Glu-Ala-Asp/His) box helicase 11 |
| chr3_+_15394428 | 0.55 |
ENSDART00000133168
|
atxn2l
|
ataxin 2-like |
| chr25_-_27665978 | 0.54 |
ENSDART00000123590
|
si:ch211-91p5.3
|
si:ch211-91p5.3 |
| chr18_-_38270596 | 0.51 |
ENSDART00000098889
|
caprin1b
|
cell cycle associated protein 1b |
| chr15_-_16384184 | 0.51 |
ENSDART00000154504
|
fam222bb
|
family with sequence similarity 222, member Bb |
| chr9_+_41459759 | 0.50 |
ENSDART00000132501
ENSDART00000100265 |
nemp2
|
nuclear envelope integral membrane protein 2 |
| chr5_+_52625975 | 0.50 |
ENSDART00000170341
ENSDART00000168317 |
apba1a
|
amyloid beta (A4) precursor protein-binding, family A, member 1a |
| chr9_+_37754845 | 0.50 |
ENSDART00000100592
|
pdia5
|
protein disulfide isomerase family A, member 5 |
| chr18_+_19990412 | 0.49 |
ENSDART00000155054
ENSDART00000090310 |
pias1b
|
protein inhibitor of activated STAT, 1b |
| chr14_-_25078569 | 0.49 |
ENSDART00000172802
ENSDART00000173345 ENSDART00000135004 |
matr3l1.1
|
matrin 3-like 1.1 |
| chr3_-_48612078 | 0.48 |
ENSDART00000169923
|
ndel1b
|
nudE neurodevelopment protein 1-like 1b |
| chr8_+_54081819 | 0.48 |
ENSDART00000005857
ENSDART00000161795 |
prickle2a
|
prickle homolog 2a |
| chr20_-_23254876 | 0.48 |
ENSDART00000141510
|
ociad1
|
OCIA domain containing 1 |
| chr16_+_26439518 | 0.48 |
ENSDART00000041787
|
trim35-28
|
tripartite motif containing 35-28 |
| chr1_-_38361496 | 0.48 |
ENSDART00000015323
|
fbxo8
|
F-box protein 8 |
| chr17_-_35076730 | 0.48 |
ENSDART00000146590
|
mboat2a
|
membrane bound O-acyltransferase domain containing 2a |
| chr22_-_5958066 | 0.47 |
ENSDART00000145821
|
si:rp71-36a1.3
|
si:rp71-36a1.3 |
| chr15_+_45591669 | 0.47 |
ENSDART00000157459
|
atg16l1
|
ATG16 autophagy related 16-like 1 (S. cerevisiae) |
| chr13_+_7578111 | 0.46 |
ENSDART00000175431
|
gbf1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
| chr11_-_30341431 | 0.46 |
ENSDART00000078378
|
cflara
|
CASP8 and FADD-like apoptosis regulator a |
| chr10_+_2899108 | 0.45 |
ENSDART00000147031
|
erap1a
|
endoplasmic reticulum aminopeptidase 1a |
| chr6_-_1566407 | 0.45 |
ENSDART00000112118
|
trim107
|
tripartite motif containing 107 |
| chr11_-_20096018 | 0.45 |
ENSDART00000030420
|
ogfrl2
|
opioid growth factor receptor-like 2 |
| chr15_-_15227541 | 0.44 |
ENSDART00000184787
|
rrp8
|
ribosomal RNA processing 8, methyltransferase, homolog (yeast) |
| chr14_+_22397251 | 0.43 |
ENSDART00000185239
ENSDART00000124072 ENSDART00000054977 |
atp7a
|
ATPase copper transporting alpha |
| chr12_+_2648043 | 0.43 |
ENSDART00000082220
|
gdf2
|
growth differentiation factor 2 |
| chr6_-_1566581 | 0.42 |
ENSDART00000192993
|
trim107
|
tripartite motif containing 107 |
| chr10_-_7347311 | 0.42 |
ENSDART00000168885
|
nrg1
|
neuregulin 1 |
| chr13_-_49144799 | 0.42 |
ENSDART00000030939
ENSDART00000148922 |
disc1
|
disrupted in schizophrenia 1 |
| chr12_-_26559650 | 0.41 |
ENSDART00000153401
|
tap2t
|
transporter associated with antigen processing, subunit type t, teleost specific |
| chr11_+_1584747 | 0.41 |
ENSDART00000154583
|
si:dkey-40c23.2
|
si:dkey-40c23.2 |
| chr11_-_45385803 | 0.41 |
ENSDART00000173329
|
trappc10
|
trafficking protein particle complex 10 |
| chr3_-_367283 | 0.41 |
ENSDART00000155936
ENSDART00000161964 ENSDART00000158560 ENSDART00000135595 ENSDART00000145890 |
mhc1zaa
|
major histocompatibility complex class I ZAA |
| chr10_-_42147318 | 0.41 |
ENSDART00000134890
|
dusp11
|
dual specificity phosphatase 11 (RNA/RNP complex 1-interacting) |
| chr5_+_45895791 | 0.40 |
ENSDART00000141039
|
polk
|
polymerase (DNA directed) kappa |
| chr20_-_3403033 | 0.40 |
ENSDART00000092264
|
rev3l
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr7_-_804515 | 0.40 |
ENSDART00000159359
|
rela
|
v-rel avian reticuloendotheliosis viral oncogene homolog A |
| chr5_-_23715027 | 0.40 |
ENSDART00000139020
|
gbgt1l1
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 1 |
| chr3_+_36127287 | 0.39 |
ENSDART00000058605
ENSDART00000182500 |
scpep1
|
serine carboxypeptidase 1 |
| chr11_-_45385602 | 0.38 |
ENSDART00000166691
|
trappc10
|
trafficking protein particle complex 10 |
| chr14_-_33083539 | 0.38 |
ENSDART00000160173
|
dlg3
|
discs, large homolog 3 (Drosophila) |
| chr20_-_38525467 | 0.38 |
ENSDART00000061417
|
si:ch211-245h14.1
|
si:ch211-245h14.1 |
| chr7_+_66884291 | 0.38 |
ENSDART00000187499
|
sbf2
|
SET binding factor 2 |
| chr19_+_7115223 | 0.37 |
ENSDART00000001359
|
psmb12
|
proteasome subunit beta 12 |
| chr1_-_524433 | 0.37 |
ENSDART00000147610
|
si:ch73-41e3.7
|
si:ch73-41e3.7 |
| chr7_+_22792895 | 0.37 |
ENSDART00000184407
|
rbm4.3
|
RNA binding motif protein 4.3 |
| chr21_+_40695345 | 0.37 |
ENSDART00000143594
|
ccdc82
|
coiled-coil domain containing 82 |
| chr17_-_28811747 | 0.36 |
ENSDART00000001444
|
g2e3
|
G2/M-phase specific E3 ubiquitin protein ligase |
| chr5_-_37820878 | 0.36 |
ENSDART00000165513
|
AL935065.1
|
Danio rerio uncharacterized LOC567472 (LOC567472), mRNA. |
| chr9_+_23765587 | 0.35 |
ENSDART00000145120
|
si:ch211-219a4.3
|
si:ch211-219a4.3 |
| chr8_-_51367298 | 0.35 |
ENSDART00000060628
|
chmp7
|
charged multivesicular body protein 7 |
| chr6_-_1566186 | 0.34 |
ENSDART00000156305
|
trim107
|
tripartite motif containing 107 |
| chr14_-_48786708 | 0.34 |
ENSDART00000169730
|
si:ch211-199b20.3
|
si:ch211-199b20.3 |
| chr5_+_51111766 | 0.34 |
ENSDART00000188552
|
pomt1
|
protein-O-mannosyltransferase 1 |
| chr22_-_8006342 | 0.34 |
ENSDART00000162028
|
sc:d217
|
sc:d217 |
| chr6_+_9793791 | 0.33 |
ENSDART00000149896
|
als2b
|
amyotrophic lateral sclerosis 2b (juvenile) |
| chr20_+_49787584 | 0.33 |
ENSDART00000193458
ENSDART00000181511 ENSDART00000185850 ENSDART00000185613 ENSDART00000191671 |
CABZ01078261.1
|
|
| chr16_+_29514473 | 0.33 |
ENSDART00000034102
|
ctss2.2
|
cathepsin S, ortholog 2, tandem duplicate 2 |
| chr7_+_54260004 | 0.31 |
ENSDART00000171909
|
pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr19_+_20178978 | 0.31 |
ENSDART00000145115
ENSDART00000151175 |
tra2a
|
transformer 2 alpha homolog |
| chr16_-_50952266 | 0.31 |
ENSDART00000165408
|
si:dkeyp-97a10.3
|
si:dkeyp-97a10.3 |
| chr22_-_10158038 | 0.31 |
ENSDART00000047444
|
rbck1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr5_-_825920 | 0.31 |
ENSDART00000126982
|
zgc:158463
|
zgc:158463 |
| chr3_+_938438 | 0.30 |
ENSDART00000165671
|
irgf4
|
immunity-related GTPase family, f4 |
| chr23_-_12906228 | 0.30 |
ENSDART00000138807
|
ndnl2
|
necdin-like 2 |
| chr7_+_66884570 | 0.30 |
ENSDART00000082664
|
sbf2
|
SET binding factor 2 |
| chr21_+_35328025 | 0.28 |
ENSDART00000136211
|
rars
|
arginyl-tRNA synthetase |
| chr19_+_42336523 | 0.28 |
ENSDART00000151304
|
anp32e
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr11_-_19775182 | 0.28 |
ENSDART00000037894
|
namptb
|
nicotinamide phosphoribosyltransferase b |
| chr18_+_50525109 | 0.27 |
ENSDART00000098390
|
ubl7b
|
ubiquitin-like 7b (bone marrow stromal cell-derived) |
| chr6_+_41554794 | 0.27 |
ENSDART00000165424
|
srgap3
|
SLIT-ROBO Rho GTPase activating protein 3 |
| chr22_-_7461603 | 0.26 |
ENSDART00000170630
|
BX511034.6
|
|
| chr9_-_9960940 | 0.26 |
ENSDART00000092164
|
prmt2
|
protein arginine methyltransferase 2 |
| chr22_+_29990448 | 0.26 |
ENSDART00000165313
|
si:dkey-286j15.3
|
si:dkey-286j15.3 |
| chr9_-_23765480 | 0.26 |
ENSDART00000027212
|
si:ch211-219a4.6
|
si:ch211-219a4.6 |
| chr5_+_13373593 | 0.25 |
ENSDART00000051668
ENSDART00000183883 |
ccl19a.2
|
chemokine (C-C motif) ligand 19a, tandem duplicate 2 |
| chr1_-_57129179 | 0.25 |
ENSDART00000157226
ENSDART00000152469 |
si:ch73-94k4.2
|
si:ch73-94k4.2 |
| chr3_-_40136743 | 0.24 |
ENSDART00000149546
|
myo15aa
|
myosin XVAa |
| chr1_-_52447364 | 0.24 |
ENSDART00000140740
|
si:ch211-217k17.10
|
si:ch211-217k17.10 |
| chr4_-_75641117 | 0.24 |
ENSDART00000182165
|
si:dkey-71l4.2
|
si:dkey-71l4.2 |
| chr8_+_47897734 | 0.24 |
ENSDART00000140266
|
mfn2
|
mitofusin 2 |
| chr15_-_8177919 | 0.23 |
ENSDART00000101463
|
bach1a
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 a |
| chr23_-_36449111 | 0.23 |
ENSDART00000110478
|
zgc:174906
|
zgc:174906 |
| chr22_+_32235922 | 0.23 |
ENSDART00000114531
ENSDART00000136125 |
rbm15b
|
RNA binding motif protein 15B |
| chr3_+_48473346 | 0.22 |
ENSDART00000166294
|
metrnl
|
meteorin, glial cell differentiation regulator-like |
| chr1_-_47089818 | 0.21 |
ENSDART00000132378
|
itsn1
|
intersectin 1 (SH3 domain protein) |
| chr5_+_32815745 | 0.21 |
ENSDART00000181535
|
crata
|
carnitine O-acetyltransferase a |
| chr9_+_34334156 | 0.21 |
ENSDART00000144272
|
pou2f1b
|
POU class 2 homeobox 1b |
| chr21_-_11632403 | 0.21 |
ENSDART00000171708
ENSDART00000138619 ENSDART00000136308 ENSDART00000144770 |
cast
|
calpastatin |
| chr5_+_45895613 | 0.21 |
ENSDART00000083937
|
polk
|
polymerase (DNA directed) kappa |
| chr6_+_42338309 | 0.20 |
ENSDART00000015277
|
gpx1b
|
glutathione peroxidase 1b |
| chr13_-_50200348 | 0.19 |
ENSDART00000038391
|
pkz
|
protein kinase containing Z-DNA binding domains |
| chr24_+_12945803 | 0.19 |
ENSDART00000005105
|
psme1
|
proteasome activator subunit 1 |
| chr6_+_52947699 | 0.19 |
ENSDART00000180913
|
uba7
|
ubiquitin-like modifier activating enzyme 7 |
| chr23_+_39970425 | 0.19 |
ENSDART00000165376
|
fyco1a
|
FYVE and coiled-coil domain containing 1a |
| chr14_+_52481288 | 0.18 |
ENSDART00000169164
ENSDART00000159297 |
tcerg1a
|
transcription elongation regulator 1a (CA150) |
| chr16_+_5901835 | 0.18 |
ENSDART00000060519
|
ulk4
|
unc-51 like kinase 4 |
| chr7_-_2309236 | 0.17 |
ENSDART00000173356
|
si:dkey-187j14.8
|
si:dkey-187j14.8 |
| chr22_-_11520405 | 0.16 |
ENSDART00000063157
|
slc26a11
|
solute carrier family 26 (anion exchanger), member 11 |
| chr3_+_19336286 | 0.16 |
ENSDART00000111528
|
kri1
|
KRI1 homolog |
| chr7_-_30492018 | 0.16 |
ENSDART00000099639
ENSDART00000162705 ENSDART00000173663 |
adam10a
|
ADAM metallopeptidase domain 10a |
| chr20_-_36408836 | 0.14 |
ENSDART00000076419
|
lbr
|
lamin B receptor |
| chr17_+_27134806 | 0.14 |
ENSDART00000151901
|
rps6ka1
|
ribosomal protein S6 kinase a, polypeptide 1 |
| chr22_+_15979430 | 0.14 |
ENSDART00000189703
ENSDART00000192674 |
rc3h1a
|
ring finger and CCCH-type domains 1a |
| chr5_+_38680061 | 0.14 |
ENSDART00000143259
|
si:dkey-58f10.11
|
si:dkey-58f10.11 |
| chr19_-_3742472 | 0.13 |
ENSDART00000162132
|
btr22
|
bloodthirsty-related gene family, member 22 |
| chr20_-_26936887 | 0.13 |
ENSDART00000160827
|
ftr79
|
finTRIM family, member 79 |
| chr6_+_52947186 | 0.13 |
ENSDART00000155831
|
uba7
|
ubiquitin-like modifier activating enzyme 7 |
| chr16_+_48631412 | 0.13 |
ENSDART00000154273
|
pbx2
|
pre-B-cell leukemia homeobox 2 |
| chr6_-_45869127 | 0.13 |
ENSDART00000062459
ENSDART00000180563 |
rbm19
|
RNA binding motif protein 19 |
| chr10_+_40623414 | 0.12 |
ENSDART00000187616
|
zgc:172131
|
zgc:172131 |
| chr5_+_36895860 | 0.12 |
ENSDART00000134493
|
srsf7a
|
serine/arginine-rich splicing factor 7a |
| chr5_+_38685089 | 0.12 |
ENSDART00000139743
|
si:dkey-58f10.10
|
si:dkey-58f10.10 |
| chr18_+_8812549 | 0.12 |
ENSDART00000017619
|
impdh1a
|
IMP (inosine 5'-monophosphate) dehydrogenase 1a |
| chr11_-_44485700 | 0.12 |
ENSDART00000186613
ENSDART00000172828 ENSDART00000113222 |
mfn1b
|
mitofusin 1b |
| chr8_+_50983551 | 0.11 |
ENSDART00000142061
|
si:dkey-32e23.4
|
si:dkey-32e23.4 |
| chr22_+_1502561 | 0.11 |
ENSDART00000159973
|
AL954741.3
|
|
| chr23_-_21763598 | 0.11 |
ENSDART00000145408
|
vps13d
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr8_-_36469117 | 0.11 |
ENSDART00000111240
|
mhc2dab
|
major histocompatibility complex class II DAB gene |
| chr16_-_11986321 | 0.10 |
ENSDART00000148666
ENSDART00000029121 |
usp5
|
ubiquitin specific peptidase 5 (isopeptidase T) |
| chr23_+_46183410 | 0.10 |
ENSDART00000167596
ENSDART00000151149 ENSDART00000150896 |
btr31
|
bloodthirsty-related gene family, member 31 |
| chr23_+_24604481 | 0.10 |
ENSDART00000146332
|
kansl2
|
KAT8 regulatory NSL complex subunit 2 |
| chr1_+_8304904 | 0.10 |
ENSDART00000168631
|
cacna1hb
|
calcium channel, voltage-dependent, T type, alpha 1H subunit b |
| chr18_-_14901437 | 0.09 |
ENSDART00000145842
ENSDART00000008035 |
trabd
|
TraB domain containing |
| chr11_+_40544269 | 0.08 |
ENSDART00000175424
ENSDART00000184673 |
mmel1
|
membrane metallo-endopeptidase-like 1 |
| chr22_+_1508545 | 0.08 |
ENSDART00000164814
|
zgc:173486
|
zgc:173486 |
| chr16_+_48678655 | 0.08 |
ENSDART00000150156
|
tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr22_-_12862415 | 0.08 |
ENSDART00000145156
ENSDART00000137280 |
glsa
|
glutaminase a |
| chr22_-_5518117 | 0.07 |
ENSDART00000164613
|
CABZ01064972.1
|
|
| chr2_+_24374669 | 0.07 |
ENSDART00000133818
|
nr2f6a
|
nuclear receptor subfamily 2, group F, member 6a |
| chr21_+_35327589 | 0.07 |
ENSDART00000145191
ENSDART00000132743 |
rars
|
arginyl-tRNA synthetase |
| chr24_+_24809877 | 0.06 |
ENSDART00000185117
|
dnajc5b
|
DnaJ (Hsp40) homolog, subfamily C, member 5 beta |
| chr3_+_8826905 | 0.06 |
ENSDART00000167474
ENSDART00000168088 |
si:dkeyp-30d6.2
|
si:dkeyp-30d6.2 |
| chr22_+_1683185 | 0.06 |
ENSDART00000158109
|
si:dkey-1b17.6
|
si:dkey-1b17.6 |
| chr5_+_61799629 | 0.06 |
ENSDART00000113508
|
hnrnpul1l
|
heterogeneous nuclear ribonucleoprotein U-like 1 like |
| chr4_+_25558849 | 0.06 |
ENSDART00000113663
ENSDART00000100755 ENSDART00000111416 ENSDART00000127840 ENSDART00000168618 ENSDART00000111820 ENSDART00000113866 ENSDART00000110107 ENSDART00000111344 ENSDART00000108548 |
zgc:195175
|
zgc:195175 |
| chr4_-_17669881 | 0.05 |
ENSDART00000066997
|
dram1
|
DNA-damage regulated autophagy modulator 1 |
| chr11_-_17713987 | 0.05 |
ENSDART00000090401
|
fam19a4b
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A4b |
| chr13_-_298454 | 0.05 |
ENSDART00000135782
|
chs1
|
chitin synthase 1 |
| chr22_-_17459587 | 0.05 |
ENSDART00000142267
|
si:ch211-197g15.10
|
si:ch211-197g15.10 |
| chr3_-_7997887 | 0.05 |
ENSDART00000172256
|
TRIM35 (1 of many)
|
si:ch211-175l6.2 |
| chr21_-_22635245 | 0.05 |
ENSDART00000115224
ENSDART00000101782 |
nectin1a
|
nectin cell adhesion molecule 1a |
| chr15_+_15786160 | 0.05 |
ENSDART00000130670
ENSDART00000090939 |
tada2a
|
transcriptional adaptor 2A |
| chr6_+_56076595 | 0.04 |
ENSDART00000169747
|
rtf2
|
replication termination factor 2 domain containing 1 |
| chr3_+_58092212 | 0.04 |
ENSDART00000156059
|
si:ch211-256e16.7
|
si:ch211-256e16.7 |
| chr3_-_10739625 | 0.04 |
ENSDART00000156144
|
zgc:112965
|
zgc:112965 |
| chr22_+_10158502 | 0.04 |
ENSDART00000005869
|
rpp14
|
ribonuclease P/MRP 14 subunit |
| chr22_+_9994833 | 0.04 |
ENSDART00000172696
ENSDART00000192101 |
BX324216.1
|
|
| chr3_-_12026741 | 0.04 |
ENSDART00000132238
|
cfap70
|
cilia and flagella associated protein 70 |
| chr19_+_7938121 | 0.03 |
ENSDART00000140053
ENSDART00000146649 |
si:dkey-266f7.5
|
si:dkey-266f7.5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of irf1a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:1901836 | regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901836) |
| 0.3 | 3.8 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.3 | 0.9 | GO:0090008 | hypoblast development(GO:0090008) |
| 0.2 | 1.0 | GO:2000660 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.2 | 0.8 | GO:0021755 | eurydendroid cell differentiation(GO:0021755) |
| 0.2 | 1.7 | GO:1901678 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.1 | 0.7 | GO:0030575 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.1 | 0.8 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 0.6 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.1 | 0.6 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.1 | 1.0 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.1 | 0.6 | GO:0006083 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.1 | 0.5 | GO:0039689 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.1 | 0.5 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.1 | 0.6 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 0.4 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.1 | 0.5 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 1.3 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 0.4 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.1 | 1.1 | GO:2000344 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.3 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.1 | 0.6 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.1 | 0.4 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.0 | 0.6 | GO:0042119 | granulocyte activation(GO:0036230) neutrophil activation(GO:0042119) |
| 0.0 | 0.2 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.5 | GO:1902041 | regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902041) negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.0 | 0.7 | GO:0090279 | regulation of calcium ion import(GO:0090279) |
| 0.0 | 0.2 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.2 | GO:0048662 | regulation of smooth muscle cell proliferation(GO:0048660) negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.0 | 0.2 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.0 | 0.2 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.8 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.6 | GO:2000134 | negative regulation of cell cycle G1/S phase transition(GO:1902807) negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.2 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 | 0.4 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.9 | GO:0009225 | nucleotide-sugar metabolic process(GO:0009225) |
| 0.0 | 0.1 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.0 | 0.9 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.3 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.4 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.9 | GO:0006289 | nucleotide-excision repair(GO:0006289) |
| 0.0 | 0.1 | GO:0002478 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) antigen processing and presentation of exogenous antigen(GO:0019884) antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.0 | 0.1 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 1.9 | GO:0017148 | negative regulation of translation(GO:0017148) |
| 0.0 | 0.5 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.7 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.0 | GO:0044650 | virion attachment to host cell(GO:0019062) adhesion of symbiont to host(GO:0044406) adhesion of symbiont to host cell(GO:0044650) |
| 0.0 | 0.4 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.0 | 0.3 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.1 | GO:0046037 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.0 | 0.3 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.3 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.4 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.1 | GO:0046349 | chitin biosynthetic process(GO:0006031) amino sugar biosynthetic process(GO:0046349) glucosamine-containing compound biosynthetic process(GO:1901073) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:1990071 | TRAPPII protein complex(GO:1990071) |
| 0.1 | 0.5 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.1 | 0.4 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.2 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 2.4 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.7 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.9 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.3 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.4 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 0.3 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.6 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.4 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.4 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.0 | 0.7 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.6 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.6 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.2 | 0.9 | GO:0004649 | poly(ADP-ribose) glycohydrolase activity(GO:0004649) |
| 0.1 | 0.7 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.1 | 0.4 | GO:0046978 | TAP1 binding(GO:0046978) |
| 0.1 | 0.3 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.1 | 3.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.6 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.1 | 1.3 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 0.4 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.1 | 0.4 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.1 | 0.4 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 0.4 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.1 | 0.9 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.1 | 1.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 1.0 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 0.9 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.1 | 0.2 | GO:0015462 | protein-transmembrane transporting ATPase activity(GO:0015462) |
| 0.1 | 0.2 | GO:1902945 | metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902945) |
| 0.1 | 0.2 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.0 | 0.3 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 1.0 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 2.0 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0046977 | peptide antigen-transporting ATPase activity(GO:0015433) peptide-transporting ATPase activity(GO:0015440) TAP binding(GO:0046977) |
| 0.0 | 0.4 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.5 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.9 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.8 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.5 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 1.2 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.7 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.3 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.7 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.4 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.1 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.7 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.5 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.5 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.4 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.4 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.6 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.2 | PID EPHB FWD PATHWAY | EPHB forward signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 0.9 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 0.7 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 1.0 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 0.4 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.5 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.4 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 1.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.6 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.7 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.6 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.4 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.1 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |