Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
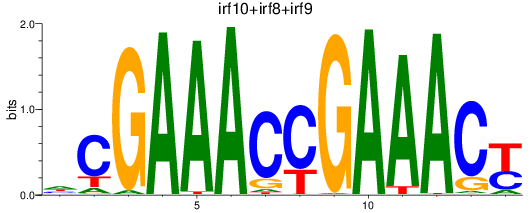
Results for irf10+irf8+irf9
Z-value: 1.38
Transcription factors associated with irf10+irf8+irf9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
irf9
|
ENSDARG00000016457 | interferon regulatory factor 9 |
|
irf10
|
ENSDARG00000027658 | interferon regulatory factor 10 |
|
irf8
|
ENSDARG00000056407 | interferon regulatory factor 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| irf8 | dr11_v1_chr18_+_30567945_30567945 | -0.83 | 5.5e-03 | Click! |
| irf9 | dr11_v1_chr12_+_13282797_13282807 | -0.63 | 6.9e-02 | Click! |
| irf10 | dr11_v1_chr23_+_642395_642395 | 0.09 | 8.2e-01 | Click! |
Activity profile of irf10+irf8+irf9 motif
Sorted Z-values of irf10+irf8+irf9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_43625549 | 1.97 |
ENSDART00000168589
|
ctsc
|
cathepsin C |
| chr9_-_35633827 | 1.78 |
ENSDART00000077745
|
zp2l1
|
zona pellucida glycoprotein 2, like 1 |
| chr22_+_25274712 | 1.53 |
ENSDART00000137341
|
BX950205.1
|
|
| chr22_+_25289360 | 1.38 |
ENSDART00000143782
|
si:ch211-154a22.8
|
si:ch211-154a22.8 |
| chr23_+_43668756 | 1.32 |
ENSDART00000112598
|
otud4
|
OTU deubiquitinase 4 |
| chr16_+_29509133 | 1.25 |
ENSDART00000112116
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr11_-_19775182 | 1.21 |
ENSDART00000037894
|
namptb
|
nicotinamide phosphoribosyltransferase b |
| chr23_-_29357764 | 1.15 |
ENSDART00000156512
|
si:ch211-129o18.4
|
si:ch211-129o18.4 |
| chr14_-_45967712 | 1.04 |
ENSDART00000043751
ENSDART00000141357 |
macrod1
|
MACRO domain containing 1 |
| chr2_+_41926707 | 0.99 |
ENSDART00000023208
|
zgc:110183
|
zgc:110183 |
| chr4_+_279669 | 0.94 |
ENSDART00000184884
|
CABZ01085275.1
|
|
| chr13_-_37631092 | 0.93 |
ENSDART00000108855
|
si:dkey-188i13.7
|
si:dkey-188i13.7 |
| chr9_+_41459759 | 0.93 |
ENSDART00000132501
ENSDART00000100265 |
nemp2
|
nuclear envelope integral membrane protein 2 |
| chr14_-_45967981 | 0.91 |
ENSDART00000188062
|
macrod1
|
MACRO domain containing 1 |
| chr13_-_37608441 | 0.88 |
ENSDART00000140230
|
zgc:123068
|
zgc:123068 |
| chr2_+_16585549 | 0.85 |
ENSDART00000134135
|
si:dkeyp-13a3.10
|
si:dkeyp-13a3.10 |
| chr3_+_36127287 | 0.83 |
ENSDART00000058605
ENSDART00000182500 |
scpep1
|
serine carboxypeptidase 1 |
| chr8_+_47188154 | 0.81 |
ENSDART00000137319
|
si:dkeyp-100a1.6
|
si:dkeyp-100a1.6 |
| chr10_+_35152928 | 0.81 |
ENSDART00000063418
|
nsun5
|
NOP2/Sun domain family, member 5 |
| chr7_+_22792895 | 0.81 |
ENSDART00000184407
|
rbm4.3
|
RNA binding motif protein 4.3 |
| chr13_+_28495419 | 0.78 |
ENSDART00000025583
|
fgf8a
|
fibroblast growth factor 8a |
| chr15_-_24588649 | 0.76 |
ENSDART00000060469
|
gemin4
|
gem (nuclear organelle) associated protein 4 |
| chr7_+_17908235 | 0.76 |
ENSDART00000077113
|
mta2
|
metastasis associated 1 family, member 2 |
| chr18_+_13248956 | 0.75 |
ENSDART00000080709
|
plcg2
|
phospholipase C, gamma 2 |
| chr15_+_24572926 | 0.75 |
ENSDART00000155636
ENSDART00000187800 |
dhrs13b
|
dehydrogenase/reductase (SDR family) member 13b |
| chr12_-_290413 | 0.74 |
ENSDART00000152496
|
adprm
|
ADP-ribose/CDP-alcohol diphosphatase, manganese-dependent |
| chr5_-_64103863 | 0.73 |
ENSDART00000135014
ENSDART00000083684 |
pappab
|
pregnancy-associated plasma protein A, pappalysin 1b |
| chr18_-_38270596 | 0.66 |
ENSDART00000098889
|
caprin1b
|
cell cycle associated protein 1b |
| chr12_-_290782 | 0.65 |
ENSDART00000152527
ENSDART00000105694 |
adprm
|
ADP-ribose/CDP-alcohol diphosphatase, manganese-dependent |
| chr19_+_7115223 | 0.64 |
ENSDART00000001359
|
psmb12
|
proteasome subunit beta 12 |
| chr18_-_38270077 | 0.63 |
ENSDART00000185546
|
caprin1b
|
cell cycle associated protein 1b |
| chr1_-_55750208 | 0.63 |
ENSDART00000142244
|
dnajb1b
|
DnaJ (Hsp40) homolog, subfamily B, member 1b |
| chr16_+_29514473 | 0.63 |
ENSDART00000034102
|
ctss2.2
|
cathepsin S, ortholog 2, tandem duplicate 2 |
| chr14_-_16810401 | 0.63 |
ENSDART00000158396
ENSDART00000170758 |
tcirg1b
|
T cell immune regulator 1, ATPase H+ transporting V0 subunit a3b |
| chr6_-_32834385 | 0.58 |
ENSDART00000129803
|
zc3h3
|
zinc finger CCCH-type containing 3 |
| chr1_-_25177086 | 0.58 |
ENSDART00000144711
ENSDART00000177225 |
tmem154
|
transmembrane protein 154 |
| chr20_+_2739804 | 0.57 |
ENSDART00000152655
|
syncrip
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr8_-_50525360 | 0.57 |
ENSDART00000175648
|
CABZ01060030.1
|
|
| chr21_+_45758498 | 0.56 |
ENSDART00000162548
|
h2afy
|
H2A histone family, member Y |
| chr4_-_17669881 | 0.56 |
ENSDART00000066997
|
dram1
|
DNA-damage regulated autophagy modulator 1 |
| chr15_-_29162193 | 0.55 |
ENSDART00000138449
ENSDART00000099885 |
xaf1
|
XIAP associated factor 1 |
| chr24_+_12945803 | 0.55 |
ENSDART00000005105
|
psme1
|
proteasome activator subunit 1 |
| chr19_+_42336523 | 0.55 |
ENSDART00000151304
|
anp32e
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr15_-_43995028 | 0.55 |
ENSDART00000172485
ENSDART00000186320 |
nlrc3l
|
NLR family, CARD domain containing 3-like |
| chr5_+_13373593 | 0.55 |
ENSDART00000051668
ENSDART00000183883 |
ccl19a.2
|
chemokine (C-C motif) ligand 19a, tandem duplicate 2 |
| chr15_-_16384184 | 0.55 |
ENSDART00000154504
|
fam222bb
|
family with sequence similarity 222, member Bb |
| chr21_+_20901505 | 0.53 |
ENSDART00000132741
|
c7b
|
complement component 7b |
| chr19_+_27448625 | 0.53 |
ENSDART00000052352
|
ppp1r11
|
protein phosphatase 1, regulatory (inhibitor) subunit 11 |
| chr17_+_25432449 | 0.53 |
ENSDART00000150195
|
srrm1
|
serine/arginine repetitive matrix 1 |
| chr18_-_38270430 | 0.52 |
ENSDART00000139519
|
caprin1b
|
cell cycle associated protein 1b |
| chr3_-_36127234 | 0.52 |
ENSDART00000130917
|
coil
|
coilin p80 |
| chr18_-_12270845 | 0.52 |
ENSDART00000143923
ENSDART00000125555 ENSDART00000062360 ENSDART00000062216 |
nup205
|
nucleoporin 205 |
| chr5_+_28497956 | 0.51 |
ENSDART00000191935
|
nfr
|
notochord formation related |
| chr22_-_17459587 | 0.51 |
ENSDART00000142267
|
si:ch211-197g15.10
|
si:ch211-197g15.10 |
| chr7_+_40081630 | 0.50 |
ENSDART00000173559
|
zgc:112356
|
zgc:112356 |
| chr10_-_42147318 | 0.49 |
ENSDART00000134890
|
dusp11
|
dual specificity phosphatase 11 (RNA/RNP complex 1-interacting) |
| chr7_-_804515 | 0.49 |
ENSDART00000159359
|
rela
|
v-rel avian reticuloendotheliosis viral oncogene homolog A |
| chr19_+_43878049 | 0.47 |
ENSDART00000165261
|
pithd1
|
PITH (C-terminal proteasome-interacting domain of thioredoxin-like) domain containing 1 |
| chr16_+_25285998 | 0.47 |
ENSDART00000154112
|
si:dkey-29h14.10
|
si:dkey-29h14.10 |
| chr17_-_35076730 | 0.47 |
ENSDART00000146590
|
mboat2a
|
membrane bound O-acyltransferase domain containing 2a |
| chr22_+_17828267 | 0.47 |
ENSDART00000136016
|
hapln4
|
hyaluronan and proteoglycan link protein 4 |
| chr5_-_39171302 | 0.47 |
ENSDART00000020808
|
paqr3a
|
progestin and adipoQ receptor family member IIIa |
| chr12_-_18578218 | 0.46 |
ENSDART00000125803
|
zdhhc4
|
zinc finger, DHHC-type containing 4 |
| chr15_-_15227541 | 0.46 |
ENSDART00000184787
|
rrp8
|
ribosomal RNA processing 8, methyltransferase, homolog (yeast) |
| chr22_-_37565348 | 0.46 |
ENSDART00000149482
ENSDART00000104478 |
fxr1
|
fragile X mental retardation, autosomal homolog 1 |
| chr5_-_67661102 | 0.45 |
ENSDART00000013605
|
zbtb20
|
zinc finger and BTB domain containing 20 |
| chr2_+_20868286 | 0.45 |
ENSDART00000062591
|
odr4
|
odr-4 GPCR localization factor homolog |
| chr12_-_18578432 | 0.45 |
ENSDART00000122858
|
zdhhc4
|
zinc finger, DHHC-type containing 4 |
| chr15_-_5624361 | 0.44 |
ENSDART00000176446
ENSDART00000114410 |
wdr62
|
WD repeat domain 62 |
| chr19_+_7597936 | 0.44 |
ENSDART00000142955
|
s100s
|
S100 calcium binding protein S |
| chr22_+_2512154 | 0.44 |
ENSDART00000097363
|
zgc:173726
|
zgc:173726 |
| chr15_+_14378829 | 0.44 |
ENSDART00000160145
|
si:ch211-11n16.2
|
si:ch211-11n16.2 |
| chr15_-_17071328 | 0.43 |
ENSDART00000122617
|
si:ch211-24o10.6
|
si:ch211-24o10.6 |
| chr15_-_37867995 | 0.43 |
ENSDART00000192698
|
si:dkey-238d18.4
|
si:dkey-238d18.4 |
| chr12_-_13905307 | 0.42 |
ENSDART00000152400
|
dbf4b
|
DBF4 zinc finger B |
| chr14_-_25078569 | 0.42 |
ENSDART00000172802
ENSDART00000173345 ENSDART00000135004 |
matr3l1.1
|
matrin 3-like 1.1 |
| chr5_+_38662125 | 0.42 |
ENSDART00000136949
|
si:dkey-58f10.13
|
si:dkey-58f10.13 |
| chr5_+_36895860 | 0.41 |
ENSDART00000134493
|
srsf7a
|
serine/arginine-rich splicing factor 7a |
| chr4_-_16658514 | 0.41 |
ENSDART00000133837
|
dennd5b
|
DENN/MADD domain containing 5B |
| chr8_+_26879358 | 0.41 |
ENSDART00000132485
|
rimkla
|
ribosomal modification protein rimK-like family member A |
| chr3_-_1938588 | 0.40 |
ENSDART00000013001
ENSDART00000186405 |
zgc:152753
|
zgc:152753 |
| chr5_-_24869213 | 0.40 |
ENSDART00000112287
ENSDART00000144635 |
gas2l1
|
growth arrest-specific 2 like 1 |
| chr19_-_7033636 | 0.40 |
ENSDART00000122815
ENSDART00000123443 ENSDART00000124094 |
daxx
|
death-domain associated protein |
| chr20_+_2739531 | 0.39 |
ENSDART00000152269
|
syncrip
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr6_+_38896158 | 0.39 |
ENSDART00000029930
ENSDART00000131347 |
slc48a1b
|
solute carrier family 48 (heme transporter), member 1b |
| chr18_+_19990412 | 0.39 |
ENSDART00000155054
ENSDART00000090310 |
pias1b
|
protein inhibitor of activated STAT, 1b |
| chr3_-_6759534 | 0.39 |
ENSDART00000180862
|
mast1b
|
microtubule associated serine/threonine kinase 1b |
| chr21_+_5192016 | 0.39 |
ENSDART00000139288
|
si:dkey-121h17.7
|
si:dkey-121h17.7 |
| chr25_-_6420800 | 0.38 |
ENSDART00000153768
|
ptpn9a
|
protein tyrosine phosphatase, non-receptor type 9, a |
| chr11_-_20096018 | 0.38 |
ENSDART00000030420
|
ogfrl2
|
opioid growth factor receptor-like 2 |
| chr9_+_23765587 | 0.38 |
ENSDART00000145120
|
si:ch211-219a4.3
|
si:ch211-219a4.3 |
| chr5_-_26247215 | 0.38 |
ENSDART00000136806
|
erap1b
|
endoplasmic reticulum aminopeptidase 1b |
| chr5_+_32845759 | 0.37 |
ENSDART00000136552
|
si:ch211-208h16.4
|
si:ch211-208h16.4 |
| chr15_+_20352123 | 0.37 |
ENSDART00000011030
ENSDART00000163532 ENSDART00000169537 ENSDART00000161047 |
il15l
|
interleukin 15, like |
| chr1_-_21563040 | 0.36 |
ENSDART00000049572
|
ncapd3
|
non-SMC condensin II complex, subunit D3 |
| chr8_-_25422186 | 0.35 |
ENSDART00000113492
ENSDART00000131736 |
kcnq2a
|
potassium voltage-gated channel, KQT-like subfamily, member 2a |
| chr4_-_12795030 | 0.34 |
ENSDART00000150427
|
b2m
|
beta-2-microglobulin |
| chr2_-_37744951 | 0.34 |
ENSDART00000144807
|
myo9b
|
myosin IXb |
| chr10_+_7719796 | 0.34 |
ENSDART00000191795
|
ggcx
|
gamma-glutamyl carboxylase |
| chr15_+_24588963 | 0.34 |
ENSDART00000155075
|
zgc:198241
|
zgc:198241 |
| chr11_+_1584747 | 0.34 |
ENSDART00000154583
|
si:dkey-40c23.2
|
si:dkey-40c23.2 |
| chr25_+_35942867 | 0.33 |
ENSDART00000066985
|
hsd17b2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr23_+_2666944 | 0.33 |
ENSDART00000192861
|
CABZ01057928.1
|
|
| chr16_+_26439939 | 0.33 |
ENSDART00000143073
|
trim35-28
|
tripartite motif containing 35-28 |
| chr5_+_45895613 | 0.32 |
ENSDART00000083937
|
polk
|
polymerase (DNA directed) kappa |
| chr18_+_26422124 | 0.32 |
ENSDART00000060245
|
ctsh
|
cathepsin H |
| chr5_-_45958838 | 0.31 |
ENSDART00000135072
|
poc5
|
POC5 centriolar protein homolog (Chlamydomonas) |
| chr22_-_10570749 | 0.30 |
ENSDART00000140736
|
si:dkey-42i9.6
|
si:dkey-42i9.6 |
| chr1_-_44928987 | 0.30 |
ENSDART00000134635
|
si:dkey-9i23.15
|
si:dkey-9i23.15 |
| chr6_-_52234796 | 0.30 |
ENSDART00000001803
|
tomm34
|
translocase of outer mitochondrial membrane 34 |
| chr19_-_27448606 | 0.30 |
ENSDART00000079251
|
vars2
|
valyl-tRNA synthetase 2, mitochondrial (putative) |
| chr23_-_27692717 | 0.30 |
ENSDART00000053878
ENSDART00000145028 |
IKZF4
|
si:dkey-166n8.9 |
| chr22_-_5958066 | 0.29 |
ENSDART00000145821
|
si:rp71-36a1.3
|
si:rp71-36a1.3 |
| chr12_+_14149686 | 0.29 |
ENSDART00000123741
|
kbtbd2
|
kelch repeat and BTB (POZ) domain containing 2 |
| chr2_-_38363017 | 0.29 |
ENSDART00000088026
|
prmt5
|
protein arginine methyltransferase 5 |
| chr9_-_23765480 | 0.29 |
ENSDART00000027212
|
si:ch211-219a4.6
|
si:ch211-219a4.6 |
| chr4_-_76383223 | 0.28 |
ENSDART00000161866
ENSDART00000168456 ENSDART00000174287 ENSDART00000174048 |
zgc:123107
|
zgc:123107 |
| chr8_-_11202378 | 0.28 |
ENSDART00000147817
ENSDART00000174039 |
fam208b
|
family with sequence similarity 208, member B |
| chr5_+_36611128 | 0.28 |
ENSDART00000097684
|
nova1
|
neuro-oncological ventral antigen 1 |
| chr4_+_40329524 | 0.28 |
ENSDART00000165722
|
znf993
|
zinc finger protein 993 |
| chr5_-_57723929 | 0.27 |
ENSDART00000144237
|
gig2p
|
grass carp reovirus (GCRV)-induced gene 2p |
| chr23_+_24955394 | 0.27 |
ENSDART00000142124
|
nol9
|
nucleolar protein 9 |
| chr14_+_21754521 | 0.27 |
ENSDART00000111839
|
kdm2ab
|
lysine (K)-specific demethylase 2Ab |
| chr15_-_1484795 | 0.27 |
ENSDART00000129356
|
si:dkeyp-97b10.3
|
si:dkeyp-97b10.3 |
| chr22_-_6941098 | 0.27 |
ENSDART00000105864
|
zgc:171500
|
zgc:171500 |
| chr16_-_5115993 | 0.26 |
ENSDART00000138654
|
ttk
|
ttk protein kinase |
| chr19_+_4066449 | 0.26 |
ENSDART00000162461
|
btr26
|
bloodthirsty-related gene family, member 26 |
| chr20_-_20402500 | 0.26 |
ENSDART00000190747
ENSDART00000185065 |
prkchb
|
protein kinase C, eta, b |
| chr21_-_30994577 | 0.25 |
ENSDART00000065503
|
pgap2
|
post-GPI attachment to proteins 2 |
| chr3_+_40164129 | 0.25 |
ENSDART00000102526
|
gfer
|
growth factor, augmenter of liver regeneration (ERV1 homolog, S. cerevisiae) |
| chr4_-_41269844 | 0.25 |
ENSDART00000186177
|
CR388165.2
|
|
| chr13_-_25548733 | 0.24 |
ENSDART00000168099
ENSDART00000135788 ENSDART00000077655 |
mcmbp
|
minichromosome maintenance complex binding protein |
| chr18_+_34667486 | 0.24 |
ENSDART00000163583
|
CR626874.1
|
|
| chr6_+_32834760 | 0.24 |
ENSDART00000121562
|
cyldl
|
cylindromatosis (turban tumor syndrome), like |
| chr5_-_30382925 | 0.23 |
ENSDART00000125381
|
gig2o
|
grass carp reovirus (GCRV)-induced gene 2o |
| chr22_-_7461603 | 0.23 |
ENSDART00000170630
|
BX511034.6
|
|
| chr18_-_12858016 | 0.23 |
ENSDART00000130343
|
parp12a
|
poly (ADP-ribose) polymerase family, member 12a |
| chr7_-_29043075 | 0.23 |
ENSDART00000052342
|
thap11
|
THAP domain containing 11 |
| chr15_-_17169935 | 0.23 |
ENSDART00000110111
|
cul5a
|
cullin 5a |
| chr5_+_45895791 | 0.22 |
ENSDART00000141039
|
polk
|
polymerase (DNA directed) kappa |
| chr3_+_938438 | 0.22 |
ENSDART00000165671
|
irgf4
|
immunity-related GTPase family, f4 |
| chr14_+_31566517 | 0.22 |
ENSDART00000002684
|
ints6l
|
integrator complex subunit 6 like |
| chr16_-_25285469 | 0.21 |
ENSDART00000183943
ENSDART00000191103 ENSDART00000154543 |
prelid3a
|
PRELI domain containing 3A |
| chr22_-_8174244 | 0.21 |
ENSDART00000156571
|
si:ch73-44m9.3
|
si:ch73-44m9.3 |
| chr8_+_1187928 | 0.21 |
ENSDART00000127252
|
slc35d2
|
solute carrier family 35 (UDP-GlcNAc/UDP-glucose transporter), member D2 |
| chr16_+_26439518 | 0.21 |
ENSDART00000041787
|
trim35-28
|
tripartite motif containing 35-28 |
| chr23_-_31974060 | 0.20 |
ENSDART00000168087
|
BX927210.1
|
|
| chr21_+_17542473 | 0.20 |
ENSDART00000005750
ENSDART00000141326 |
stom
|
stomatin |
| chr10_+_7255695 | 0.20 |
ENSDART00000161506
ENSDART00000181304 |
fut10
|
fucosyltransferase 10 |
| chr15_+_2421729 | 0.20 |
ENSDART00000082294
ENSDART00000156428 |
hephl1a
|
hephaestin-like 1a |
| chr6_-_52235118 | 0.20 |
ENSDART00000191243
|
tomm34
|
translocase of outer mitochondrial membrane 34 |
| chr3_+_24603923 | 0.19 |
ENSDART00000172589
|
si:dkey-68o6.6
|
si:dkey-68o6.6 |
| chr8_+_22277198 | 0.19 |
ENSDART00000005989
|
dffb
|
DNA fragmentation factor, beta polypeptide (caspase-activated DNase) |
| chr22_-_17474583 | 0.18 |
ENSDART00000148027
|
si:ch211-197g15.8
|
si:ch211-197g15.8 |
| chr12_-_19151708 | 0.18 |
ENSDART00000057124
|
tefa
|
thyrotrophic embryonic factor a |
| chr21_-_22648007 | 0.18 |
ENSDART00000121788
|
gig2l
|
grass carp reovirus (GCRV)-induced gene 2l |
| chr12_-_26559650 | 0.18 |
ENSDART00000153401
|
tap2t
|
transporter associated with antigen processing, subunit type t, teleost specific |
| chr12_-_35936329 | 0.18 |
ENSDART00000166634
|
rnf213b
|
ring finger protein 213b |
| chr2_-_40889465 | 0.18 |
ENSDART00000192631
ENSDART00000180824 |
uggt1
|
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr25_+_32390794 | 0.18 |
ENSDART00000012600
|
galk2
|
galactokinase 2 |
| chr22_+_1589407 | 0.17 |
ENSDART00000175704
|
si:ch211-255f4.11
|
si:ch211-255f4.11 |
| chr3_+_52999962 | 0.17 |
ENSDART00000104683
|
pbx4
|
pre-B-cell leukemia transcription factor 4 |
| chr19_+_22085925 | 0.17 |
ENSDART00000185636
|
atp9b
|
ATPase phospholipid transporting 9B |
| chr10_+_26645953 | 0.16 |
ENSDART00000131482
|
adgrg4b
|
adhesion G protein-coupled receptor G4b |
| chr3_-_52661242 | 0.16 |
ENSDART00000138018
|
zgc:113210
|
zgc:113210 |
| chr23_+_29357790 | 0.16 |
ENSDART00000139223
ENSDART00000141928 |
tardbpl
|
TAR DNA binding protein, like |
| chr10_+_31809226 | 0.16 |
ENSDART00000087898
|
foxo1b
|
forkhead box O1 b |
| chr7_-_52417060 | 0.16 |
ENSDART00000148579
|
myzap
|
myocardial zonula adherens protein |
| chr11_-_31276064 | 0.16 |
ENSDART00000141062
ENSDART00000004780 |
man2b1
|
mannosidase, alpha, class 2B, member 1 |
| chr22_+_8979955 | 0.16 |
ENSDART00000144005
|
si:ch211-213a13.1
|
si:ch211-213a13.1 |
| chr3_-_44753325 | 0.15 |
ENSDART00000074543
|
hs3st4
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 4 |
| chr19_+_7124337 | 0.15 |
ENSDART00000031380
|
tap2a
|
transporter associated with antigen processing, subunit type a |
| chr4_-_26095755 | 0.15 |
ENSDART00000100611
ENSDART00000191266 |
si:ch211-244b2.3
|
si:ch211-244b2.3 |
| chr25_-_27665978 | 0.15 |
ENSDART00000123590
|
si:ch211-91p5.3
|
si:ch211-91p5.3 |
| chr7_+_56651759 | 0.15 |
ENSDART00000073600
|
kcng4b
|
potassium voltage-gated channel, subfamily G, member 4b |
| chr14_-_26465729 | 0.14 |
ENSDART00000143454
|
syvn1
|
synovial apoptosis inhibitor 1, synoviolin |
| chr16_-_11986321 | 0.14 |
ENSDART00000148666
ENSDART00000029121 |
usp5
|
ubiquitin specific peptidase 5 (isopeptidase T) |
| chr4_-_58945951 | 0.14 |
ENSDART00000159145
|
si:dkey-28i19.4
|
si:dkey-28i19.4 |
| chr3_-_4455951 | 0.14 |
ENSDART00000193908
ENSDART00000074077 |
trim35-3
|
tripartite motif containing 35-3 |
| chr1_+_38858399 | 0.14 |
ENSDART00000165454
|
CU915762.1
|
|
| chr6_+_52947699 | 0.14 |
ENSDART00000180913
|
uba7
|
ubiquitin-like modifier activating enzyme 7 |
| chr21_-_40557281 | 0.14 |
ENSDART00000172327
|
taok1b
|
TAO kinase 1b |
| chr22_+_2459591 | 0.14 |
ENSDART00000182602
|
si:ch73-92e7.4
|
si:ch73-92e7.4 |
| chr25_-_35524166 | 0.13 |
ENSDART00000156782
|
slc5a12
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 12 |
| chr1_-_50710468 | 0.13 |
ENSDART00000080389
|
fam13a
|
family with sequence similarity 13, member A |
| chr13_-_37642890 | 0.13 |
ENSDART00000146483
ENSDART00000136071 ENSDART00000111786 |
si:dkey-188i13.9
|
si:dkey-188i13.9 |
| chr3_-_45361573 | 0.13 |
ENSDART00000154796
|
il21r.1
|
interleukin 21 receptor, tandem duplicate 1 |
| chr6_+_32834007 | 0.13 |
ENSDART00000157353
|
cyldl
|
cylindromatosis (turban tumor syndrome), like |
| chr3_-_12026741 | 0.13 |
ENSDART00000132238
|
cfap70
|
cilia and flagella associated protein 70 |
| chr15_-_15965456 | 0.13 |
ENSDART00000154579
|
synrg
|
synergin, gamma |
| chr6_-_43270160 | 0.12 |
ENSDART00000180990
|
frmd4ba
|
FERM domain containing 4Ba |
| chr4_+_35198656 | 0.12 |
ENSDART00000187990
|
si:dkey-269p2.1
|
si:dkey-269p2.1 |
| chr1_-_7951002 | 0.12 |
ENSDART00000138187
|
si:dkey-79f11.8
|
si:dkey-79f11.8 |
| chr13_+_7578111 | 0.12 |
ENSDART00000175431
|
gbf1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
| chr22_+_1541083 | 0.12 |
ENSDART00000169338
|
si:ch211-255f4.7
|
si:ch211-255f4.7 |
| chr3_-_7997887 | 0.12 |
ENSDART00000172256
|
TRIM35 (1 of many)
|
si:ch211-175l6.2 |
| chr2_-_32688905 | 0.12 |
ENSDART00000041146
|
nrbp2a
|
nuclear receptor binding protein 2a |
| chr3_-_32079916 | 0.11 |
ENSDART00000040900
|
baxb
|
BCL2 associated X, apoptosis regulator b |
| chr22_-_17474781 | 0.11 |
ENSDART00000186817
|
si:ch211-197g15.8
|
si:ch211-197g15.8 |
| chr4_-_75616197 | 0.11 |
ENSDART00000157778
|
si:dkey-71l4.3
|
si:dkey-71l4.3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of irf10+irf8+irf9
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:2000660 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.1 | 0.8 | GO:0061193 | rhombomere 5 development(GO:0021571) rhombomere 6 development(GO:0021572) tongue development(GO:0043586) tongue morphogenesis(GO:0043587) taste bud development(GO:0061193) taste bud morphogenesis(GO:0061194) taste bud formation(GO:0061195) |
| 0.1 | 1.0 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 | 0.5 | GO:0030575 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.1 | 0.5 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.1 | 0.4 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.1 | 0.3 | GO:1902102 | meiotic cell cycle phase transition(GO:0044771) metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.1 | 0.8 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.1 | 0.5 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.1 | 0.3 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 0.1 | 0.3 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.1 | 0.5 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 | 0.5 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.1 | 0.3 | GO:0071480 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.1 | 0.4 | GO:2000105 | positive regulation of DNA-dependent DNA replication(GO:2000105) |
| 0.1 | 0.3 | GO:0014005 | microglia development(GO:0014005) |
| 0.1 | 0.3 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.1 | GO:0055057 | neuroblast division(GO:0055057) |
| 0.0 | 0.3 | GO:0002504 | antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.0 | 0.6 | GO:0007035 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.0 | 0.5 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.4 | GO:1901678 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.0 | 0.2 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.3 | GO:0034122 | negative regulation of toll-like receptor signaling pathway(GO:0034122) |
| 0.0 | 0.2 | GO:0003232 | bulbus arteriosus development(GO:0003232) negative regulation of histone methylation(GO:0031061) |
| 0.0 | 0.4 | GO:0042119 | granulocyte activation(GO:0036230) neutrophil activation(GO:0042119) |
| 0.0 | 0.2 | GO:0030262 | apoptotic DNA fragmentation(GO:0006309) cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.0 | 0.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.1 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.0 | 0.2 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.9 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.8 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.1 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.5 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.1 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.6 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.4 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.5 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.1 | GO:0031179 | peptide modification(GO:0031179) |
| 0.0 | 0.1 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.0 | 0.5 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.2 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.7 | GO:0009395 | phospholipid catabolic process(GO:0009395) |
| 0.0 | 1.7 | GO:0017148 | negative regulation of translation(GO:0017148) |
| 0.0 | 0.1 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.1 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.4 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.4 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.5 | GO:2000045 | regulation of G1/S transition of mitotic cell cycle(GO:2000045) |
| 0.0 | 0.5 | GO:0006626 | protein targeting to mitochondrion(GO:0006626) |
| 0.0 | 0.2 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 0.4 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.8 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.3 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.4 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.1 | GO:0001836 | release of cytochrome c from mitochondria(GO:0001836) |
| 0.0 | 0.2 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 0.5 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 0.5 | GO:1902737 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.1 | 1.0 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 0.8 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 0.5 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 0.2 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 0.6 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 0.3 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.5 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.3 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.8 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.3 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.3 | GO:0042611 | MHC protein complex(GO:0042611) MHC class II protein complex(GO:0042613) |
| 0.0 | 0.5 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.5 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.1 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.1 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 0.9 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.3 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 5.1 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 1.6 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.4 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 0.4 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) citrate-L-glutamate ligase activity(GO:0072591) |
| 0.1 | 1.7 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 0.5 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.1 | 1.4 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 0.5 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.1 | 0.8 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 0.3 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.1 | 0.6 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 1.0 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 0.3 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.1 | 0.3 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.1 | 0.3 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.1 | 0.3 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.1 | 0.2 | GO:0046978 | TAP1 binding(GO:0046978) |
| 0.1 | 0.5 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.2 | GO:0005461 | UDP-glucuronic acid transmembrane transporter activity(GO:0005461) UDP-N-acetylgalactosamine transmembrane transporter activity(GO:0005463) |
| 0.0 | 0.4 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.6 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.6 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.2 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.0 | 0.1 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 0.2 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.4 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.3 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.8 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.1 | GO:0004649 | poly(ADP-ribose) glycohydrolase activity(GO:0004649) |
| 0.0 | 0.4 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.7 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.1 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.9 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.0 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.2 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0036374 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 2.5 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.4 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.5 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.0 | 0.4 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.5 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.4 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.5 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.0 | 0.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.5 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.3 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 1.0 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.6 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 0.5 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.3 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.7 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 1.6 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.5 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 1.5 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.4 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.2 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |